Scientists at the Institut Pasteur and McMaster University have discovered that the evolution of a gene in the bacterium that causes bubonic plague, Yersinia pestis, may have prolonged the duration of two major pandemics. They have demonstrated that modifying the copy number of a specific virulence gene increases the length of infection in affected individuals. It is thought that this genetic change may prompt longer periods of contagiousness in less densely populated environments, in which the time of transmission from one individual to another is inevitably longer. This genetic variation has been observed in strains of each of the two major plague pandemics, hundreds of years before they eventually faded out. The study was published in the journal Science on May 29, 2025.
There are three major plague pandemics documented in human history. The first broke out in the Mediterranean basin in the 6th century. The second emerged in the 14th century, reemerging on several occasions in Europe over a period of more than 500 years. The first wave of this second pandemic, known as the "Black Death," remains the deadliest event ever recorded in human history, killing an estimated 30 to 50 per cent of the European population between 1347 and 1352. The third plague pandemic occurred in Asia in 1850. This pandemic spread to and across every continent, and still persists today in "endemic" regions, encompassing countries like Uganda and the Democratic Republic of Congo, as well as the United States and Mongolia.
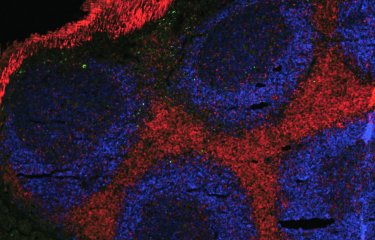
The plague bacillus, or Y. pestis, remains highly fatal due to the presence of various factors that drive its virulence, including the gene known as pla, a high copy component in the genome of the Y. pestis bacterium. This virulence factor enables the bacterium to travel to the lymph nodes and multiply there before spreading to the rest of the body, causing rapid septicemia.
By studying hundreds of samples from ancient plague victims, the researchers at McMaster University, who specialize in ancient DNA, observed a decrease in the copy number of the gene pla in the later stages of both the first and second pandemics. To back up this observation, the Institut Pasteur scientists studied the third plague pandemic, testing living contemporary strains from samples preserved in a collection at the institute.
"Ours is one of the first research studies to directly examine changes in an ancient pathogen, one we still see today, in an attempt to understand what drives the virulence, persistence, and eventual extinction of pandemics," says Hendrik Poinar, co-senior author of the study, director of the McMaster Ancient DNA Centre, and holder of the Michael G. DeGroote Chair in Genetic Anthropology.
The National Reference Center for Plague and Yersinioses at the Institut Pasteur houses one of the world’s richest collections of modern Y. pestis isolates. "Thanks to our international collaborators who monitor local epidemics of plague worldwide, we were able to find the unique bacterial samples used for this project," says Javier Pizarro-Cerdá, co-senior author of the work and director of the Yersinia Research Unit and of the WHO Collaborating Center for Plague at the Institut Pasteur. "We identified three samples of Y. pestis found in Asia in the 1990s in which the total number of pla genes had decreased," says Guillem Mas Fiol, co-lead author of the study and researcher in the Yersinia unit at the Institut Pasteur. "These three samples enabled us to analyze the biological impact of these pla gene deletions in vitro and in vivo, thereby corroborating our findings with the paleogenomic observations made by our Canadian colleagues," adds Javier Pizarro-Cerdá.
In mice models of bubonic plague, the scientists found that depletion in the copy number of the pla gene leads to a 20 per cent decrease in mortality and an increase in the length of infection in affected individuals, meaning the infected rodents lived longer before they died.
The scientists propose that rats infected with these bacteria could spread infection farther in an environment with a reduced mammal population density. The high mortality rate among rodents at the start of pandemic waves drives a decrease in host proximity. "Diminished virulence may give the bacillus a selective advantage within a reduced population density," explains Javier Pizarro-Cerdá. The pandemics eventually faded out, probably due to the reduced virulence of these strains.
This genetic evolution happened randomly and independently in each historical plague pandemic. "Our research sheds light on an interesting pattern in the evolutionary history of the plague. However, it is important to note that the majority of strains which continue to circulate today in Africa, the Americas, and Asia are highly virulent strains, the ones that were previously responsible for mass mortality," says Ravneet Sidhu, co-lead author of the study, and PhD student at the Ancient DNA Centre at McMaster University. The deadly effects of infection with the plague bacillus, Y. pestis, are now more controlled thanks to antibiotics and diagnostic methods, which have changed the evolutionary dynamics.
"Today, the plague is a rare disease, but one that remains a public health concern and serves as a model for gaining a broad understanding of how pandemics emerge and become extinct. This example illustrates the balance of virulence a pathogen can adopt in order to spread effectively from one host to another," concludes Javier Pizarro-Cerdá.
Source
Attenuation of virulence in Yersinia pestis across three plague pandemics, Science, May 29, 2025
Ravneet Kaur Sidhu*1,2,†, Guillem Mas Fiol3,4,†, Pierre Lê-Bury3,4,5, Christian E. Demeure4, Emelyne Bougit3, Rémi Beau3, Charlotte Balière6, Aurelia Kwasiborski6, Valérie Caro6, Jennifer Klunk1,7, Daniel J. Salkeld8, Ann Carmichael9, Nükhet Varlık10, Debi Poinar1, David J.D. Earn11,14, Benjamin M. Bolker2,11, Jonathan Dushoff2,14, G. Brian Golding2, Nicolas Rascovan12, Olivier Dussurget3,4, Edward C. Holmes13, Javier Pizarro-Cerdá*3,4 and Hendrik N. Poinar*1,14, 15
1McMaster Ancient DNA Centre, Departments of Anthropology and Biochemistry, McMaster University, Hamilton, Ontario, Canada L8S4L9.
2Department of Biology, McMaster University, Hamilton, Ontario, Canada L8S4L9.
3Institut Pasteur, Université Paris-Cité, Yersinia Research Unit, 7-5015 Paris, France.
4Institut Pasteur, Université Paris Cité, WHO Collaborating Research and Reference Centre for Plague FRA-146, F-75015 Paris, France
5Center for Immunology of Viral, Auto-immune, Hematological and Bacterial Diseases (IMVA- HB/IDMIT), Université Paris-Saclay, Inserm, CEA, F-92260 Fontenay-aux-Roses, France 6Institut Pasteur, Université Paris-Cité, Environment and Infectious Risk Unit, Laboratory for Urgent Response to Biological Threats, F-75015, Paris, France
7Daciel Arbor Biosciences, Ann Arbor, MI, USA
8Department of Biology, Colorado State University, CO, USA.
9History Department, Indiana University, Bloomington, IN, USA.
10Department of History, Rutgers University-Newark, NJ, USA.
11Department of Mathematics and Statistics, McMaster University, Hamilton, Ontario, Canada L8S4K1.
12Institut Pasteur, Université Paris-Cité, Microbial Palogenomics Unit, F-75015, Paris, France 13School of Medical Sciences, University of Sydney, Sydney, NSW 2006. Australia.
14Michael G. DeGroote Institute of Infectious Disease Research, McMaster University, Hamilton, Ontario, Canada L8S4L9.
15Canadian Institute for Advanced Research, Toronto, Canada.
†These authors contributed equally to this work