Using the 2005-2006 chikungunya virus epidemic that occurred in the Indian Ocean islands as a study model, researchers at the Pasteur Institute have succeeded in developing an approach that can predict the virus mutations most likely to emerge in the short-term, with strong epidemic potential. This work has strong implications for improving surveillance of ongoing epidemics and the potential to strengthen vaccine strategies against emerging viral diseases. The results were published in the journal Cell Host & Microbe.
Press release
Paris, June 12th, 2014
The molecular mechanisms of replication and multiplication of RNA viruses are burdened by error - mutations - in their genetic code, that lead to the production of new viruses, each one slightly different from each other. These are frequent events, and so a viral population from a single strain is actually composed of a cloud of variants, with diverse mutations. Among these mutations, some will confer a selective advantage to a given variant in the population: in the case of arboviruses (such as dengue, chikungunya, West Nile, yellow fever) that cycle between insects and vertebrates, for example, they improve a virus' ability to replicate in mosquitoes and transmit to mammals, rendering it more likely to spread than others. The new strain in question thus bears a stronger likelihood of causing a new epidemic.
Reseachers in Marco Vignuzzi's unit (Institut Pasteur/CNRS) have succeeded for the first time in developing a method to predict the mutations most likely to emerge within an arbovirus population, with strong epidemic potential. Their method relies on monitoring the natural, rather than forced, evolution of a virus during a transmission cycle between mosquitoes and mammals.
In a first step, the researchers validated their approach in a proof-of-concept by addressing the emergence event that led to the chikungunya epidemic of 2005-2006 that occurred in the Indian Ocean. To do so, they naturally infected two species of mosquitoes, Aedes aegypti (the usual vector for chikungunya) and Aedes albopictus (the Asian tiger mosquito), with the pre-epidemic strain. Next, they let the infection develop in mosquitoes for a relatively long period (14 days), corresponding to several replication cycles, and proceeded to deep sequence the virus in different anatomical sites of individual mosquitoes. The results revealed the emergence and rise to dominance, specifically in the saliva compartment, of the same mutation in the E1 glycoprotein (A226V) that was at the origin of the 2005 epidemic. Importantly, they were able to identify in a single infection cycle, what required years to emerge in nature.
The scientists then asked whether this approach could simulate the natural evolution of the post-epidemic strain towards a future epidemic. They thus repeated experiments using the strain that emerged after the 2005 epidemic, in a natural transmission study between an infected mosquito and a mouse, and then from the mouse to yet another mosquito. By the time the infection of the first mosquito developed, two new mutations (V80I and A129V, also in the E1 glycoprotein) appeared in saliva and were transmitted to mice, remaining as a minority component of the virus population. However, in the second batch of mosquitoes that fed on infected mice, the virus strains carrying these two new mutations supplanted all other variants, including the currently circulating strain, revealing the strong potential for these mutated strains to emerge after a single transmission cycle. In vitro studies suggest that the selective advantage of these mutations is linked to an increase ability to fuse and enter cells during infection and a higher virus particle stability when the virus is outside the host cell (such as in mammalian blood or mosquito saliva).
In addition to identifying and characterizing these two new mutations in the context of the Indian Ocean strain of chikungunya, this work is an innovative tool to identify new variants with epidemic potential, well ahead of when they could be identified in nature, for any arbovirus as well as for other RNA viruses for which transmission samples can be collected. This discovery now offers the possibility of improving and better targeting surveillance between and during epidemics. For viruses for which vaccines exist, such an approach could help better choose what strains to include in vaccine preparations.
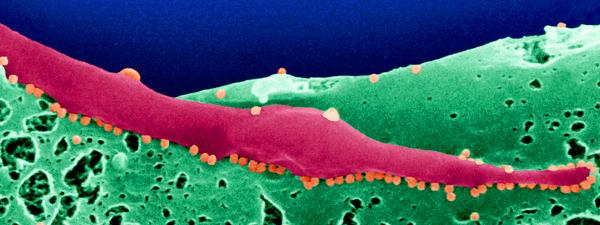
Picture: Chikungunya virus (orange) budding at the surface of infected Aedes albopictus cells. © Institut Pasteur
This study was funded by the Bill et Melinda Gates foundation, the European Research Council, the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence (grant ANR-10-LABX-62-IBEID), and the Franco-Israëli high council for science and technology.
Source
Emergence and Transmission of Arbovirus Evolutionary Intermediates with Epidemic Potential, Cell Host & Microbe, June 11th, 2014.
Kenneth A. Stapleford (1), Lark L. Coffey (2), Sreyrath Lay (3), Antonio V. Bordería (1), Veasna Duong (3), Ofer Isakov (4), Kathryn Rozen-Gagnon (1), Camilo Arias-Goeta (5), Hervé Blanc (1), Stéphanie Beaucourt (1), Türkan Haliloğlu (6), Christine Schmitt (7), Isabelle Bonne (7), Nir Ben-Tal (8), Noam Shomron (4), Anna-Bella Failloux (5), Philippe Buchy (3) and Marco Vignuzzi (1).
(1) Unité Populations virales et pathogénèse, Institut Pasteur, Centre National de la Recherche Scientifique UMR 3569, 28 rue du Dr Roux,75724 Paris Cedex 15, France
(2) Center for Vectorborne Diseases, Department of Pathology, Microbiology and Immunology, School of Veterinary Medicine, University of California, Davis, One Shields Avenue, 5327 VM3A, Davis, CA 95616, USA
(3) Unité de Virologie, Institut Pasteur in Cambodia 5, Monivong Boulevard, PO Box 983, Phnom Penh, Cambodia
(4) Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv 69978, Israel
(5) Laboratoire Arbovirus et insectes vecteurs, Institut Pasteur, 25 rue du Dr Roux, 75724 Paris Cedex 15, France
(6) Department of Chemical Engineering and Polymer Research Center, Boğaziçi University, Bebek 34342, Istanbul, Turkey
(7) Ultrastructural Microscopy Platform, Institut Pasteur, 28 rue du Dr Roux, 75724 Paris Cedex 15, France
(8) Department of Biochemistry and Molecular Biology, George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv 69978, Israel