Scientists have combined metabolic analysis, state-of-the-art microscopic techniques and artificial intelligence to successfully predict the outcome of infection with Legionella pneumophila, which causes legionellosis, at the single-cell level. This method could be applied to other pathogenic bacteria in future.
How does legionellosis develop in the early stages of infection? Which of the body's cells enable Legionella pneumophila to develop, and why? Scientists at the Institut Pasteur set out to find answers to these questions.
Legionella pneumophila bacteria are responsible for most cases of legionellosis, a group of infections including Legionnaires' disease and the milder Pontiac fever. Legionellosis is generally caused by inhaling contaminated aerosols (water droplets suspended in the air). Several cases of Legionnaires' disease are reported each year in France, and a significant outbreak recently occurred in the town of Albertville, in Savoie. Legionnaires' disease affects the lungs and may prove fatal in the most severe cases.
Which macrophages allow the bacteria to proliferate?
Legionella pneumophila bacteria grow in the body by hijacking part of the immune system. The bacteria are capable of infecting macrophages, immune cells which generally recognize and ingest pathogens as soon as they invade tissues. But some bacteria, like Legionella pneumophila, have developed strategies to evade macrophages and exploit them as replicative niches. Instead of eliminating the pathogenic bacteria, the macrophages are turned into a microbe production plant!
"Although the bacteria infect thousands of macrophages, only some of these macrophages let the bacteria proliferate," explains Pedro Escoll Guerrero, a scientist in the Biology of Intracellular Bacteria Unit at the Institut Pasteur.
The challenge is to identify which macrophages allow the proliferation of Legionella pneumophila. In a recently published study, the scientists combined several approaches to shed light on the question, including cell metabolism analysis, confocal microscopy techniques and artificial intelligence.
Mitochondria, key elements in predicting outcomes for infected cells
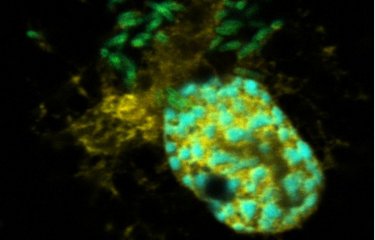
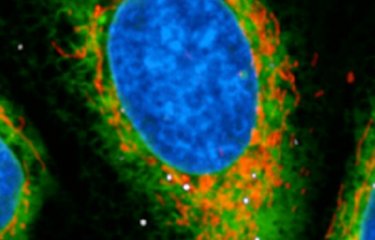
Tens of thousands of cells were infected with Legionella pneumophila and filmed in real time to determine which of them enabled the bacteria to replicate and which did not. At the same time, the scientists measured various parameters in the mitochondria of the infected cells. Previous research had demonstrated that the bacteria manipulate mitochondrial function in infected cells and inhibit the onset of cell death to support intracellular replication. So the scientists quantified mitochondrial membrane potential, in other words the difference in electrical potential between the matrix and the intermembrane space in mitochondria, and the level of mROS, molecules generated by mitochondria which play a key role in cell metabolism.
"What we discovered is that very soon after infection, the macrophages that retain high mitochondrial membrane potential and produce more mROS are precisely those that allow bacterial replication," says Pedro Escoll Guerrero.
The scientists had to process a huge quantity of metabolic measurements and images, organizing them into chronological datasets for each cell that could be used to train an artificial intelligence system, specifically a machine-learning model. The model built by the scientists based on the trajectories of more than 5,000 macrophages was then tested on more than 1,700 cells. The results showed that, based on mitochondrial parameters measured immediately after infection, we can predict with 83% accuracy whether or not Legionella pneumophila will be able to replicate in the cell.
A method applicable to other pathogenic bacteria
What next? The first step is to optimize the machine-learning model to obtain even more reliable predictions. Research is already under way in this area. The scientists then want to apply the same method to other intracellular bacteria which, like Legionella pneumophila, also infect macrophages. These include Salmonella enterica, which causes Salmonella infection, and Mycobacterium tuberculosis, the pathogen responsible for tuberculosis.
Finally, the research demonstrates that high-throughput image analysis, combined with artificial intelligence, can be used to study the heterogeneity of infection responses at the single-cell level.
Source: Backtracking metabolic dynamics in single cells predicts bacterial replication in human macrophages, Nature communications, October 16, 2025
Mariatou Dramé1,4, Francisco-Javier Garcia-Rodriguez1,4, Dmitry Ershov2,3, Jessica E. Martyn1, Jean-Yves Tinevez2, Carmen Buchrieser1,5 & Pedro Escoll1,5
1Institut Pasteur, Université Paris Cité, Biologie des Bactéries Intracellulaires, Paris, France.
2Institut Pasteur, Université Paris Cité, Image Analysis Hub, Paris, France.
3Institut Pasteur, Université Paris Cité, Bioinformatics and Biostatistics Hub, Paris, France.
4These authors contributed equally: Mariatou Dramé, Francisco-Javier Garcia-Rodriguez.
5Corresponding authors : Carmen Buchrieser, Pedro Escoll