Scientists from the Institut Pasteur have retrospectively identified early cases of Salmonella resistance to ampicillin, a broad-spectrum antibiotic that is still widely used today. By analyzing the genomes of hundreds of historical samples of Salmonella, they proved that resistance first emerged well before ampicillin was released on the market for human use. Their discovery suggests that low doses of penicillin G, routinely fed to livestock in North America and Europe in the 1950s to boost growth, may have encouraged bacteria resistant to this new antibiotic to evolve and spread to humans just a few years later. The findings were published in the journal The Lancet Infectious Diseases on November 29.
Every year, bacterial resistance to antibiotics leads to almost 25,000 deaths in Europe. Estimates suggest that by 2050, more than 10 million people a year will die as a result of antibiotic resistance worldwide. Many bacteria that cause serious infections in humans, such as Salmonella, have already developed resistance to commonly used antibiotics.
Ampicillin was the first synthetic penicillin found to be effective in treating Enterobacteriaceae infections.1 This broad-spectrum antibiotic, one of the most widespread antibiotics in the world, was released on European markets in 1961. Shortly afterwards (1962-1964), the first outbreaks caused by ampicillin-resistant strains were observed in humans. These outbreaks, caused by the zoonotic bacterium Salmonella typhimurium, particularly affected the United Kingdom.
The very short lapse of time between the release of this antibiotic on the market and the first cases of resistance described in the scientific literature prompted the Institut Pasteur's scientists to investigate the emergence of ampicillin resistance. They discovered that bacteria capable of passing on genes with ampicillin resistance actually emerged, unexpectedly, in the late 1950s.
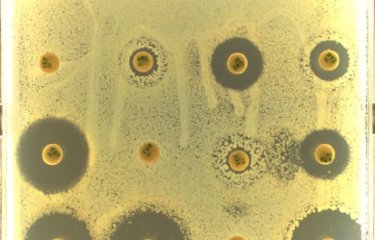
The scientists examined 288 historical samples of S. typhimurium isolated from humans, animals and food over four continents between 1911 and 1969. The samples were tested for antibiotic susceptibility and whole genome sequencing was performed to identify ampicillin resistance mechanisms and establish links between resistant strains.
The researchers found several ampicillin-resistance genes in 11 isolates (3.8%) from human samples. The blaTEM-1 gene was found on plasmids2 in three samples isolated in France and Tunisia in 1959 and 1960, several years before ampicillin was released on the market for human use.
The scientists observed that despite the close proximity between the countries, the plasmids found in the strains from France differed from those in the strains responsible for the first outbreaks in the UK in the 1960s. "This indicates that the early emergence of ampicillin resistance was caused by multiple independent acquisitions of these resistant genes by different bacterial populations, probably in response to high antibiotic pressure," explains Alicia Tran Dien, a PhD student and lead author of the paper.
The study suggests that resistance to ampicillin may have emerged as a result of the use of penicillin G3 in farms. The scientists demonstrated that ampicillin-resistance genes can be transferred between wild-type strains of S. typhimurium after exposure to relatively low levels of penicillin G, similar to the residual doses found in farms several decades ago.
"Our findings suggest that antibiotic residues in farming environments such as manure, soil and waste water in the 1950s may have had a much greater impact on the spread of ampicillin resistance than previously thought," says Dr. Francois-Xavier Weill, Head of the Institut Pasteur's Enteric Bacterial Pathogens Unit, who led the study.
"Although our study was not designed to identify a causal link between the use of penicillin G in livestock and the emergence of ampicillin resistance, the results suggest that the non-medical use of these early penicillins may have encouraged the spread of genes resistant to a new antibiotic that was effective in humans. There is therefore an urgent need to re-evaluate the use of antibiotics in animals, as the European Union has been doing over the past two decades, and to implement a "one health" approach to tackling resistance, recognizing that bacteria know no borders. This must include close surveillance of bacterial resistance in both humans and animals at international level," adds Dr. Weill.
This study comes just weeks after WHO4 called for an end to routine antibiotic use to promote growth and prevent disease in healthy farm animals.
Notes :
1 Enterobacteriaceae are a large family of Gram-negative bacteria that includes Salmonella, Escherichia coli, Yersinia pestis, Klebsiella and Shigella.
2 Plasmids are mobile DNA elements that can be easily copied and transferred between different bacteria.
3 Penicillin G is a narrow-spectrum antibiotic discovered by Alexander Fleming en 1928 but only mass-produced during the Second World War. In the 1950s it was used in North America and Europe as a growth promoter – it was added to livestock feed in low doses on a long-term basis with the aim of boosting growth and increasing market value.
4 Website :
http://who.int/mediacentre/news/releases/2017/antibiotics-animals-effectiveness/en/
Funding:
The study was funded by the Institut Pasteur, Santé publique France, the French government's Investissement d'Avenir programme (LabEX IBEID), and the Fondation Le Roch-Les Mousquetaires.
Picture : Salmonella typhimurium with peritrichous flagella (cilia). Bacterial species that causes foodborne infections. Colored image.
Source
Early transmissible ampicillin resistance in zoonotic Salmonella enterica serotype Typhimurium in the late 1950s: a retrospective, whole-genome sequencing study, The Lancet Infectious Diseases, 29 novembre 2017
Alicia Tran-Dien (a), Simon Le Hello (a), Christiane Bouchier (a), François-Xavier Weill (a)
(a) Institut Pasteur
DOI : https://doi.org/10.1016/S1473-3099(17)30705-3