In association with Imperial College London, scientists at the Institut Pasteur and CNRS have identified a vulnerable site on the surface of the dengue virus which is targeted by the only broadly neutralizing antibodies identified to date. This discovery offers a new target for the development of a vaccine to combat all four types of dengue virus currently in circulation. These results were published in the journal Nature on January 12, 2015.
Press release
Paris, January 14, 2015
Dengue is transmitted by mosquitoes, and is the most widespread viral disease in our planet’s tropical regions. Two and a half billion people live in high-risk areas. There are four strains of dengue virus, known as serotypes DEN-1, DEN-2, DEN-3 and DEN-4. A patient produces antibodies specific to one serotype during an initial infection, but these do not confer effective protection against subsequent infection by the other serotypes. These antibodies may even constitute a risk factor for developing a severe form of dengue. Any future vaccine against dengue must therefore provide effective, simultaneous protection against all four virus serotypes.
In an article published in Nature, co-signed by teams at the Institut Pasteur, CNRS and Imperial College London, scientists have highlighted the existence of a region in the virus envelope protein exposed to the antibodies that is common to all four dengue serotypes. This region would represent a promising target for the development of an effective vaccine against all four dengue serotypes.
These results were obtained in collaboration with a team from Imperial College London, who had already succeeded in identifying and isolating antibodies that simultaneously neutralize all four virus serotypes in a cohort of patients infected in Thailand. However, the mode of action of these antibodies remained unknown.
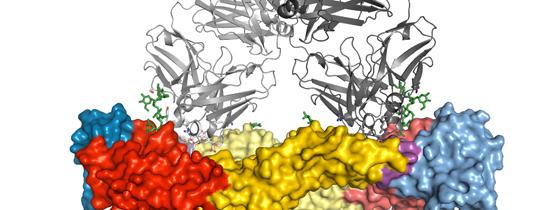
Using the SOLEIL and ESRF synchrotron radiation facilities in Saint-Aubin, near Paris and Grenoble respectively, the scientists carried out a crystallographic analysis to study the dengue virus envelope protein in complex with these specific antibodies in order to determine the 3D structure. Their analyses resulted in identification of the antibody binding site on the envelope protein. The amino acids present at this site are the same in all four virus serotypes, and therefore give it obvious potential as a vaccine target.
The authors of this study also discovered that the antibody binding site on the envelope protein doubles as the binding site for the viral protein prM. Interaction between the envelope protein and this prM protein is crucial for the production of infectious viral particles during replication of the virus in infected cells.
These observations gave the scientists an insight into the reason why this common binding site has been highly conserved in all four serotypes, despite the viral flaw that it represents: a slight mutation on this site would enable the virus to evade the immune system but, at the same time, prevents the interaction with prM that is essential for replication.
The analysis of the structures obtained using x-ray crystallography reveals that this site may be the “Achilles heel” of the dengue virus, opening up new possibilities for scientists in terms of the development of a unique antigenic determinant which would mimic this site. This antigen would potentially be able to trigger an immune response targeting all four dengue serotypes simultaneously, and could constitute a prime vaccine candidate against dengue.
Illustration: 3D structure of the antibody complexes for all four serotypes bound to the dengue virus envelope protein. © Institut Pasteur
Sources
Recognition determinants of broadly neutralizing human antibodies against dengue viruses, Nature, January 12, 2015. doi:10.1038/nature14130
Alexander Rouvinski (1,2*), Pablo Guardado-Calvo (1,2*), Giovanna Barba-Spaeth (1,2*), Stéphane Duquerroy (1,2,3), Marie-Christine Vaney (1,2), Carlos M. Kikuti (1,2), M. Erika Navarro Sanchez (1,2), Wanwisa Dejnirattisai (4), Wiyada Wongwiwat (4), Ahmed Haouz (5), Christine Girard-Blanc (5), Stéphane Petres (5), William E. Shepard (6), Philippe Desprès (7), Fernando Arenzana-Seisdedos (8), Philippe Dussart (9), Juthathip Mongkolsapaya (4,10), Gavin R. Screaton (4) & Félix A. Rey (1,2,5)
(1) Institut Pasteur, Département de Virologie, Unité de Virologie Structurale, 75724 Paris Cedex 15, France.
(2) CNRS UMR 3569 Virologie, 75724 Paris Cedex 15, France.
(3) Université Paris-Sud, Faculté des Sciences, 91405 Orsay, France.
(4) Division of Immunology and Inflammation, Department of Medicine, Hammersmith Hospital Campus, Imperial College London, London W12 0NN, UK.
(5) Institut Pasteur, Protéopôle, CNRS UMR 3528, 75724 Paris Cedex 15, France.
(6) Synchrotron SOLEIL, L’Orme des Merisiers, Saint Aubin, BP48, 91192 Gif-sur-Yvette, France.
(7) Institut Pasteur, Département de Virologie, Unité des Interactions Moléculaires Flavivirus-Hôtes, 75724 Paris Cedex 15, France.
(8) Institut Pasteur, Département de Virologie, Unité de Pathogénie Virale, INSERMU1108, 75724 Paris Cedex 15, France.
(9) Institut Pasteur de Guyane, BP 6010, 97306 Cayenne, French Guiana.
(10) Dengue Hemorrhagic Fever Research Unit, Office for Research and Development, Faculty of Medicine, Siriraj Hospital, Mahidol University, Bangkok 10700, Thailand.
* These authors contributed equally to the work.
Recognition determinants of broadly neutralizing human antibodies against dengue viruses, Nature, January 12, 2015. doi: 10.1038/nature14130
Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney MC, Kikuti CM, Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A, Girard-Blanc C, Petres S, Shepard WE, Desprès P, Arenzana-Seisdedos F, Dussart P, Mongkolsapaya J, Screaton GR, Rey FA.
A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus, Nature Immunology, December 15, 2014. doi: 10.1038/ni.3058.
Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinsky A, Jumnainsong A, Edwards C, Quyen NT, Duangchinda T, Grimes JM, Tsai W, Lai C, Wang W, Malasit P, Farrar J, Simmons CP, Zhou ZH, Rey FA, Mongkolsapaya J, Screaton GR.