Scientists from the Institut Pasteur and the CNRS, in collaboration with teams from the CNRS/Inserm/Paris Descartes University–Sorbonne Paris Cité (based at the Institut Cochin) and the Wellcome Trust (Sanger Institute), have recently revealed the cause behind the emergence in the 1960s of neonatal infections due to group B streptococcus. These findings, published in Nature Communications on August 4, 2014, prove that the sudden emergence of infections caused by this bacterium resulted from the widespread use of an antibiotic, tetracycline, from the 1950s onwards.
Press release
Paris, August 4th, 2014
Group B streptococcus (Streptococcus agalactiae or GBS) causes severe infections in about one in every two thousand newborns. The main reservoir for GBS is the gastrointestinal tract, from which it colonizes the genitourinary tract. During childbirth, pathogenic GBS bacteria may be passed on from mother to baby. Preventing neonatal GBS infection involves testing women towards the end of their pregnancy to diagnose whether they carry the bacterium. Those who test positive are given a preventive antibiotic treatment during labor.
In the 1960s, an increase in neonatal GBS infections was recorded in maternity wards in the United States and Europe. The relative absence of GBS infections in previous years may have resulted from under-diagnosis or under-reporting, but this sudden increase may have signaled a newly emerging disease whose causes were unknown.
Scientists from the Institut Pasteur/CNRS team directed by Philippe Glaser, in collaboration with teams from the CNRS/Inserm/Paris Descartes University–Sorbonne Paris Cité (based at the Institut Cochin), scientists from the Paris Public Hospital Network, the Wellcome Trust (Sanger Institute), and microbiologists from Europe and Australia, have shed new light on this mystery by tracing the evolutionary history of human group B streptococci. They sequenced and compared the genomes of 230 strains dating from the 1950s to the present day to determine their evolutionary relationships. The scientists observed that the group B streptococci population that colonizes and infects humans is composed of a small number of clones with very low genetic diversity, suggesting a recent common origin that points towards a true emergence of GBS neonatal infections.
Remarkably, some 90% of the GBS strains isolated in humans are resistant to tetracycline. By performing a genomic analysis of the clones identified by the evolutionary study, the scientists determined the genetic basis for this resistance and demonstrated that each clone was selected for its resistance to tetracycline. This observation enabled them to show the emergence of the clones as occurring in the mid-20th century, shortly after the introduction and widespread administration of one of the first antibiotics, tetracycline, as a preventive and curative measure to treat various infections in the 1950s. Based on their findings, the scientists suggested that the use of this antibiotic led to a susceptible and relatively harmless population of group B streptococci being replaced by a small number of resistant clones, which spread throughout the world. These clones, which also seem to have been selected for their potential to spread and colonize, are responsible for the infections that have been observed since the 1960s.
These findings explain the emergence of GBS infections in humans and demonstrate the long-term adverse effects of the widespread and uncontrolled use of antibiotics. Tetracycline, which was never actually used to treat group B streptococcal infection and has only rarely been prescribed over the last 20 years, had an irreversible impact on this bacterial population, currently one of the leading causes of infection in newborns. With multidrug resistance in bacteria becoming a critical issue, this study shows how important it is when prescribing any antibiotic to take into account both the direct risk of increasing multidrug resistance and also the impact on the bacterial populations that make up the natural flora in humans and animals.
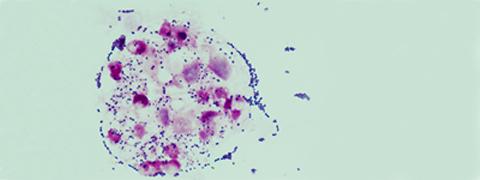
Illustration: Streptococcus agalactiae in white blood cells (polynuclear neutrophils) in the cerebrospinal fluid of a child with meningitis (optical microscopy, Gram stain). © Claire Poyart, CNR-Strep (AP-HP, Inserm)
Source
Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline, Nature Communications, August 1st, 2014, DOI: http://dx.doi.org/ 10.1038/ncomms5544
(1,2,3‡) Violette Da Cunha, (4,5,‡) Mark R. Davies, (1,2) Pierre-Emmanuel Douarre, (1,2) Isabelle Rosinski-Chupin, (6) Immaculada Margarit, (7) Sebastien Spinali, (6) Tim Perkins, (3) Pierre Lechat, (7) Nicolas Dmytruk, (1,2) Elisabeth Sauvage, (8) Laurence Ma, (6) Benedetta Romi, (8) Magali Tichit, (1,2) Maria-José Lopez-Sanchez, (3) Stéphane Descorps-Declere, (3) Erika Souche, (2,9) Carmen Buchrieser, (1,10) Patrick Trieu-Cuot, (3) Ivan Moszer, (11) Dominique Clermont, (6) Domenico Maione, (8) Christiane Bouchier, (12, 13) David J. McMillan, (4) Julian Parkhill, (6) John L. Telford, (4) Gordan Dougan, (5) Mark J. Walker, #The DEVANI Consortium, (4) Matthew T. G. Holden, (1, 7,14,15) Claire Poyart and (1,2,3 *) Philippe Glaser
(1) Institut Pasteur, Unité de Biologie des Bactéries Pathogènes à Gram-positif, Paris, France;
(2) CNRS UMR3525, Paris, France;
(3) Institut Pasteur, Bioinformatics platform, Paris, France;
(4) The Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK;
(5) Australian Infectious Diseases Research Centre, School of Chemistry and Molecular Biosciences, University of Queensland, Australia;
(6) Novartis Vaccines and Diagnostics, Siena, Italy;
(7) Centre National de Référence des Streptocoques, Groupe Hospitalier Paris Centre Cochin - Hôtel Dieu-Broca, Paris, France;
(8) Institut Pasteur Genomic platform, Paris, France;
(9) Institut Pasteur, Biologie des Bactéries Intracellulaires, Paris, France;
(10) CNRS ERL3526, Paris, France;
(11) Institut Pasteur, Collection de l'Institut Pasteur (CIP), Paris, France;
(12) QIMR Berghofer Medical Research Institute, Brisbane, Australia;
(13) Inflammation and Healing Research Cluster, University of the Sunshine Coast, Sippy Downs, Australia;
(14) Institut Cochin, Université Sorbonne Paris Descartes;
(15) INSERM, U1016, Paris, France.
‡These authors contributed equally to this work