A destabilizing factor such as a change in diet can disrupt the entire gut microbiota, with possible health consequences. An international study led by the Molecular Microbial Pathogenesis Unit (Institut Pasteur/Inserm), directed by Philippe Sansonetti, has recently demonstrated in mice that a high-fat diet has a direct influence on the gut microbiota and its environment. Bacterial communities react to this new diet with a massive reorganization, and the small intestine itself undergoes changes in its defense capacity – from the very first month. These results were published in the journal PNAS on September 16.
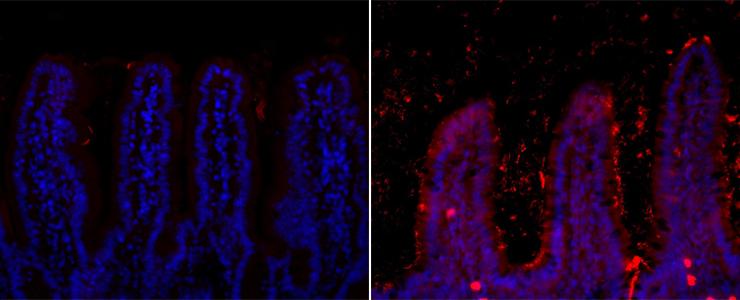
Localization of bacteria in the ileum of mice given a normal diet (left-hand image) and a high-fat diet (right-hand image). © Institut Pasteur
The billions of bacteria that make up our intestinal flora, known as the microbiota, play a vital role in digestion and also in some diseases such as type 2 diabetes and obesity. These diseases have often been linked to an imbalance in the gut microbiota, with certain bacteria becoming clearly predominant, and to increased intestinal permeability ("leaky gut syndrome"), in which inflammatory substances can be released into the bloodstream. But although there is a large body of research focusing on the state of the gut microbiota after the onset of disease, very few studies have looked at how this intestinal imbalance actually emerges, for example following the introduction of a diet that is rich in fat. An international research team therefore decided to investigate this question. "We wanted to examine how gut bacteria behave in the early stages after the introduction of a high-fat diet," explained Thierry Pédron, a research engineer in the Molecular Microbial Pathogenesis Unit (Institut Pasteur/Inserm). "We soon focused our research on the small intestine, as that was where we observed the most striking variations."
Some mice in the study were given an ordinary diet, while others were given a diet composed of 70% fat. Using genomics techniques, the scientists were able to identify the different bacterial species in feces samples and see how the composition of the microbiota changed over time. They also localized and precisely identified the bacteria within the small intestine. "Just one month after the mice started the new high-fat diet, we noticed changes in the composition of the microbiota," said Thierry Pédron. "Some bacterial species thrived while others declined. The species Candidatus arthromitus even completely disappeared. We also observed an entirely new phenomenon, namely a significant build-up of bacteria between the villi in the intestinal epithelium."
Normally bacteria are unable to approach or cross the intestinal wall, since the epithelium releases antimicrobial peptides and is lined with a protective mucus. The scientists decided to take a closer look at these defenders of the intestinal wall: they observed a significant fall in the production of antimicrobial peptides following the ingestion of large quantities of fat and a thinning of the mucus layer. In other words, fat does not only lead to the reorganization of the microbiota, it also brings about changes in the intestine itself. Further changes were also observed: additional investigations demonstrated an increase in the permeability of the small intestine and a reduction in the activity of PPARγ. "PPARγ is a molecule with many functions; it plays an important role in the metabolism of fatty acids and also in inflammation and embryogenesis," explained Thierry Pédron. "This drop in activity seems to be closely linked with the reduction in antimicrobial peptides." While the links between all these results and their potential implications in some nutritional imbalances are not yet clear, it is reassuring to note that when the mice were given a balanced diet again, everything went back to normal within a month!
Source
High-fat diet modifies the PPAR-γ pathway leading to disruption of microbial and physiological ecosystem in murine small intestine, PNAS, September 16, 2016. DOI : 10.1073/pnas.1612559113
Julie Tomasa,b,c, Céline Muleta, Azadeh Saffariana, Jean-Baptiste Cavind, Robert Ducrocd, Béatrice Regnaulte, Chek Kun Tanf, Kalina Duszkaf, Rémy Burceling,h, Walter Wahlif,i, Philippe J. Sansonettia,j,1, and Thierry Pédrona
a Unité de Pathogénie Microbienne Moléculaire, INSERM Unit U1202, Institut Pasteur, 75724 Paris Cedex 15, France;
b Institut National de la Recherche Agronomique, UMR 1319 MICALIS, F-78350 Jouy-en-Josas, France;
c AgroParisTech, UMR 1319 MICALIS, F-78350 Jouy-en-Josas, France;
d INSERM UMRS 1149, Centre de Recherche sur l’inflammation, Unité de Formation et de Recherche de Medecine Paris Diderot, F-75018 Paris, France;
e Plate-forme de Génotypage des Eucaryotes, Biomics Pole, Centre d’Innovation et Recherche Technologique, Institut Pasteur, Paris F-75015, France;
f Lee Kong Chian School of Medicine,Nanyang Technological University, Singapore;
g Institut des Maladies Métaboliques et Cardiovasculaires, INSERM U1048 F-31432 Toulouse, France;
h Université Paul Sabatier, F-31432 Toulouse, France;
i Center for Integrative Genomics, University of Lausanne, 1015 Lausanne, Switzerland; and
j Chaire de Microbiologie et Maladies Infectieuses, Collège de France, 75005 Paris, France