Rift Valley fever virus, transmitted by mosquitoes, is responsible for outbreaks in livestock in Africa and can also be fatal in humans. Scientists from the Institut Pasteur and the CNRS, working with the University of Göttingen, have characterized the mechanism used by the virus to insert one of its envelope proteins into the host cell membrane, thereby enabling it to infect the cell. They have demonstrated that the viral envelope protein has a "pocket" that specifically recognizes a category of lipids in the cell membrane. This pocket is also found in other human pathogenic viruses transmitted by different mosquitoes, such as the Zika and chikungunya viruses. Understanding these interactions should pave the way for the identification of new therapeutic strategies that target viruses transmitted by these mosquitoes. The findings were published in the journal Science on November 3, 2017.
Rift Valley fever (RVF) virus is a bunyavirus that was first isolated from sheep in Kenya in 1930. The spread of RVF has serious economic consequences in Africa. The virus also causes severe disease in humans who come into contact with contaminated animals or who are bitten by infected mosquitoes, resulting in severe encephalitis and hemorrhagic fever that can prove fatal. RVF therefore also represents a significant public health threat. In 2000, the virus spread outside the African continent to Saudi Arabia and Yemen. There are concerns that it may also extend to Asia and Europe.
RVF virus spreads in its host by fusing with cell membranes so that it can proliferate and infect other cells. Scientists in the Structural Virology Unit (Institut Pasteur/CNRS) directed by Félix Rey, in collaboration with the University of Göttingen, characterized the mechanism used by the virus to insert one of its surface proteins into the host cell membrane and drive fusion. They also determined the atomic structure of this new protein-lipid complex, demonstrating that this protein has a "pocket" which specifically recognizes the hydrophilic heads of some of the lipids that make up the cell membrane. Importantly, this "recognition pocket" is found not only in RVF virus but also in the envelope proteins of other viral families transmitted by arthropods, such as the dengue, Zika and chikungunya viruses, which have caused major worldwide epidemics in recent years. In the homologous protein of the chikungunya virus, the scientists pinpointed one of the residues of the recognition pocket as amino acid 226. In 2006, the A226V mutation enabled chikungunya to be transmitted by a new species of mosquito that is prevalent on Reunion Island (Aedes albopictus, or the tiger mosquito).
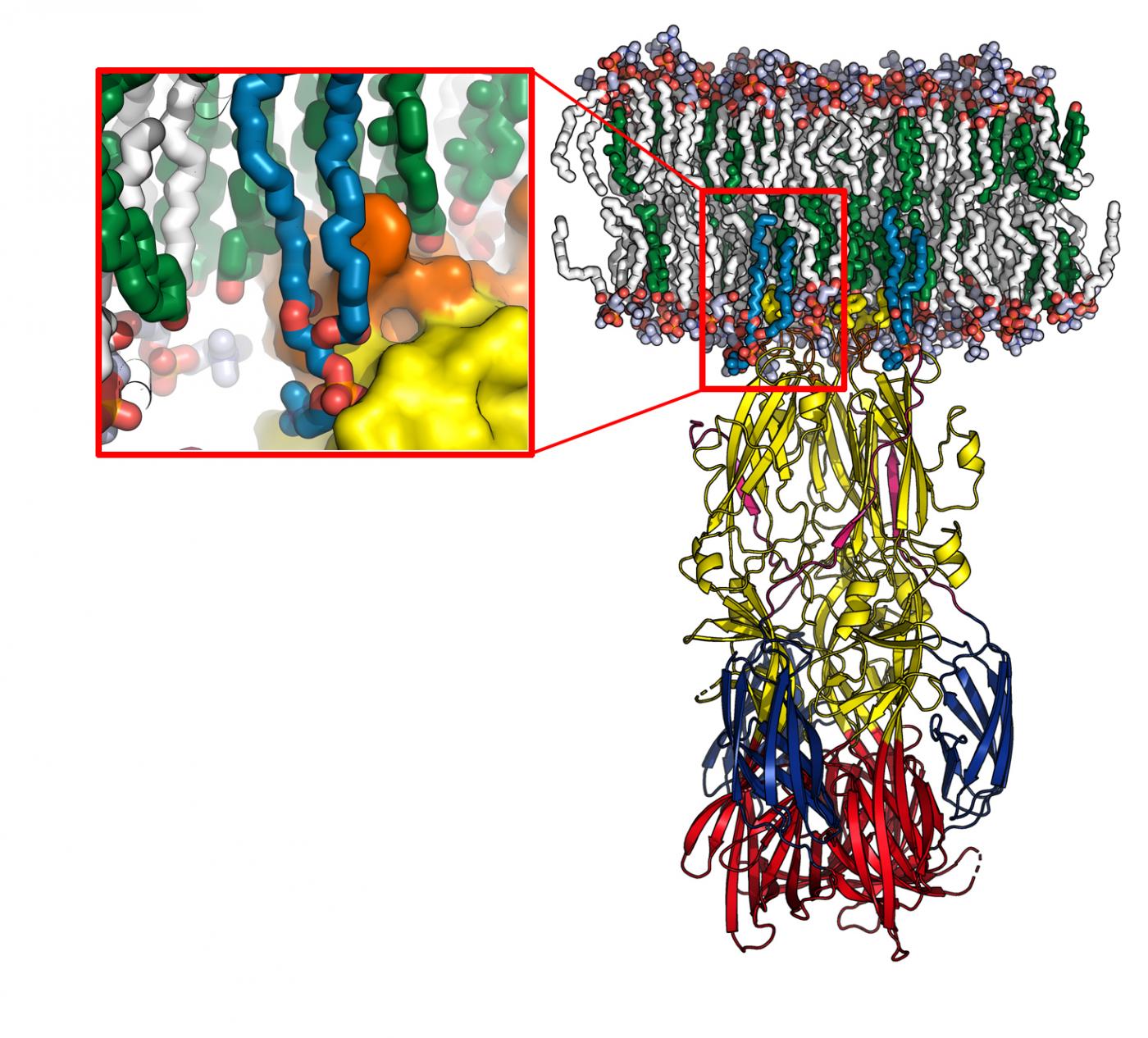
Model of the fusion protein interacting with a cell membrane (in white and green), obtained using molecular dynamics. The enlargement shows the lipid head "recognition pocket", with the protein represented at the surface. © Pablo Guardado-Calvo, Institut Pasteur
"This study offers a further illustration of the power of comparative analyses of viruses that appear very distant, such as bunyaviruses, alphaviruses and flaviviruses, which can result in highly significant findings and reveal shared mechanisms of action," commented Félix Rey, Head of the Structural Virology Unit (Institut Pasteur/CNRS), where the study was carried out.
Understanding the mechanism used by these viruses for insertion in the cell membrane paves the way for the development of therapeutic agents that target the "pocket" involved in the fusion of viral and cell membranes with the aim of preventing pathogenic arboviruses from entering host cells.
Source
* A glycerophospholipid-specific pocket in the RVFV class II fusion protein drives target membrane insertion, Science, November 3, 2017
P. Guardado-Calvo,1,2 K. Atkovska,3 S. A. Jeffers,1,2† N. Grau,1,2‡ M. Backovic,1,2 J. Pérez-Vargas,1,2§ S. M. de Boer,4 M. A. Tortorici,1,2 G. Pehau-Arnaudet,5 J. Lepault,6 P. England,5,7 P. J. Rottier,4 B. J. Bosch,4 J. S. Hub,3 F. A. Rey,1,2
(1) Institut Pasteur, Département de Virologie, Unité de Virologie Structurale, 75724 Paris Cedex 15, France.
(2) UMR 3569 Virologie, CNRS–Institut Pasteur, 25–28 Rue du Docteur Roux, 75015 Paris, France
(3) Institute for Microbiology and Genetics, University of Goettingen, Justus-von-Liebig weg 11, 37077 Göttingen, Germany
(4) Department of Infectious Diseases and 4Immunology, Virology Division, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands
(5) UMR 3528, CNRS, Institut Pasteur, 25-28 rue du Docteur Roux, 75015 Paris, France
(6) UMR9198, Institut de biologie intégrative de la cellule, CNRS, CEA, Université Paris Sud, France
(7) Institut Pasteur, Proteopole, Plateforme de Biophysique des Macromolécules et de leurs Interactions (PFBMI), 25-28 rue du Dr Roux, F-75724 Paris Cedex 15, France