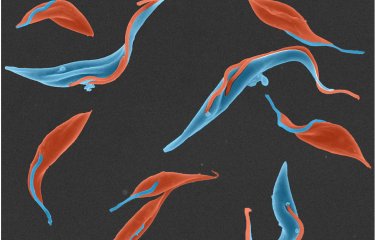
A new CRISPR-based RNA test detects animal trypanosomes, which helps track parasites that pose a threat to livestock in Africa and monitor the transmission of human-infectious species. This study was conducted by an international consortium led by the Institut Pasteur. Its outcomes are wholly aligned with both the Institut Pasteur's long-standing mission of fighting infectious disease threats and the World Health Organization's goal of eliminating human African trypanosomiasis by 2030.
Human African trypanosomiasis, commonly known as sleeping sickness, is characterized by symptoms ranging from severe fatigue and skin signs to sleep impairment and neurological disorders. The disease is caused by the parasitic flagellate Trypanosoma brucei gambiense, which is injected into the body by the tsetse fly. The insect itself becomes infected by biting infected humans or domesticated animals such as pigs.
|
A course about Neglected Tropical Diseases on FUN MOOC platform We recommend taking the "Neglected Tropical Diseases (NTDs)" MOOC, which looks at a broad and representative range of NTDs including human African trypanosomiasis. The course covers patient diagnosis, treatment and management, as well as pathogenicity mechanisms. Since most of these diseases are zoonoses, they are addressed from a "One Health" perspective, encompassing specific veterinary, entomological and epidemiological issues. |
This disease presents a major public health issue, particularly in Central Africa, where it has still not been fully eradicated in numerous foci. While sleeping sickness now only affects around 500 people per year, some trypanosomes threaten farmers' financial security, as they also cause sickness in carrier livestock. The estimated annual cost of animal infections for Africa as a whole is $4 billion.
SHERLOCK4HAT: an RNA test allowing ultra-sensitive and accurate targeted detection
Details of this discovery, coordinated by Brice Rotureau, Research Director at the Institut Pasteur in Paris and Head of the Parasitology Unit at the Institut Pasteur de Guinée, and his team, were published in the Elife journal on September 23, 2025. The work ties in with Priority 1 of the Institut Pasteur (Paris) strategic plan to tackle infectious disease threats, a mission also pursued by the Pasteur Network, to which the Institut Pasteur in Paris and the Institut Pasteur de Guinée both belong. The project's innovation lies in a brand-new molecular test that accurately identifies parasite species living in an animal's blood. Data from reservoir animals, and sentinel animals in particular, can then be used to prevent transmission to humans.
"Thanks to SHERLOCK, we can now accurately detect the main trypanosome species circulating in highly exposed livestock farming settings for the first time," explains Brice Rotureau. This new method was developed in collaboration with the team led by Lucy Glover, Research Director at the Institut Pasteur in Paris. It involves actively detecting an invariable region of trypanosomes' genetic material specific to each species. This remarkably sensitive and specific system is capable of identifying various parasite species living in farm animals.
Its aim as a surveillance tool is to break human-infectious trypanosomes' chain of infection. By improving animal diagnostics, it could be possible to limit the transmission of the parasite to humans through targeted treatment of infected animals.
A collaborative, surveillance-based approach
In addition to the technological innovation described above, the study conducted in Guinea and Côte d'Ivoire revealed that, in West Africa, farm pigs act as reservoirs for human-infectious trypanosomes. Based on numbers of individual animals carrying the parasite, decisions can be made as to whether or not alerts should be issued, and the data can also be used to guide prevention policies. This approach combines animal screening with vector control measures involving local communities. Test results are used to guide the distribution of tsetse fly traps, which are then set up by residents to limit the risk of bites.
"The collaborative dimension of this project was one of its real strengths. This firstly entailed setting up an international consortium including teams in Paris (Institut Pasteur), Montpellier (IRD), Guinea and Côte d’Ivoire. We also combined laboratory work with direct applications in the field, encouraging long-term engagement from local populations," says Brice Rotureau. This collective approach helps map infections more accurately and encourages preventive measures aimed at driving progress on the strategy to achieve "zero transmission" of human African trypanosomiasis by 2030.
Expanding the scope of surveillance and developing applications for other diseases
This RNA test paves the way for new applications in addition to monitoring the transmission of various trypanosome species. It could also soon be used to assess parasites' resistance to treatments, an increasingly urgent issue given the emergence of multi-drug-resistant parasite strains. While surveillance is very useful for effective response and efforts to eradicate human cases, there are limits to its benefits. "To address challenges faced in the field, we aim to adapt this test so it can be used quickly and directly at the point of care. Moreover, the benefits of this technology could be rolled out to other parasitic diseases such as Chagas disease and leishmaniasis," concludes Brice Rotureau.
Source
Roger-Junior Eloiflin, Elena Pérez-Antón ... Brice Rotureau. A SHERLOCK toolbox for the eco-epidemiological surveillance of animal African trypanosomosis reveals a similar parasite diversity in domestic pigs in two ancient sleeping sickness foci in Western Africa. eLife. Sept 22, 2025.