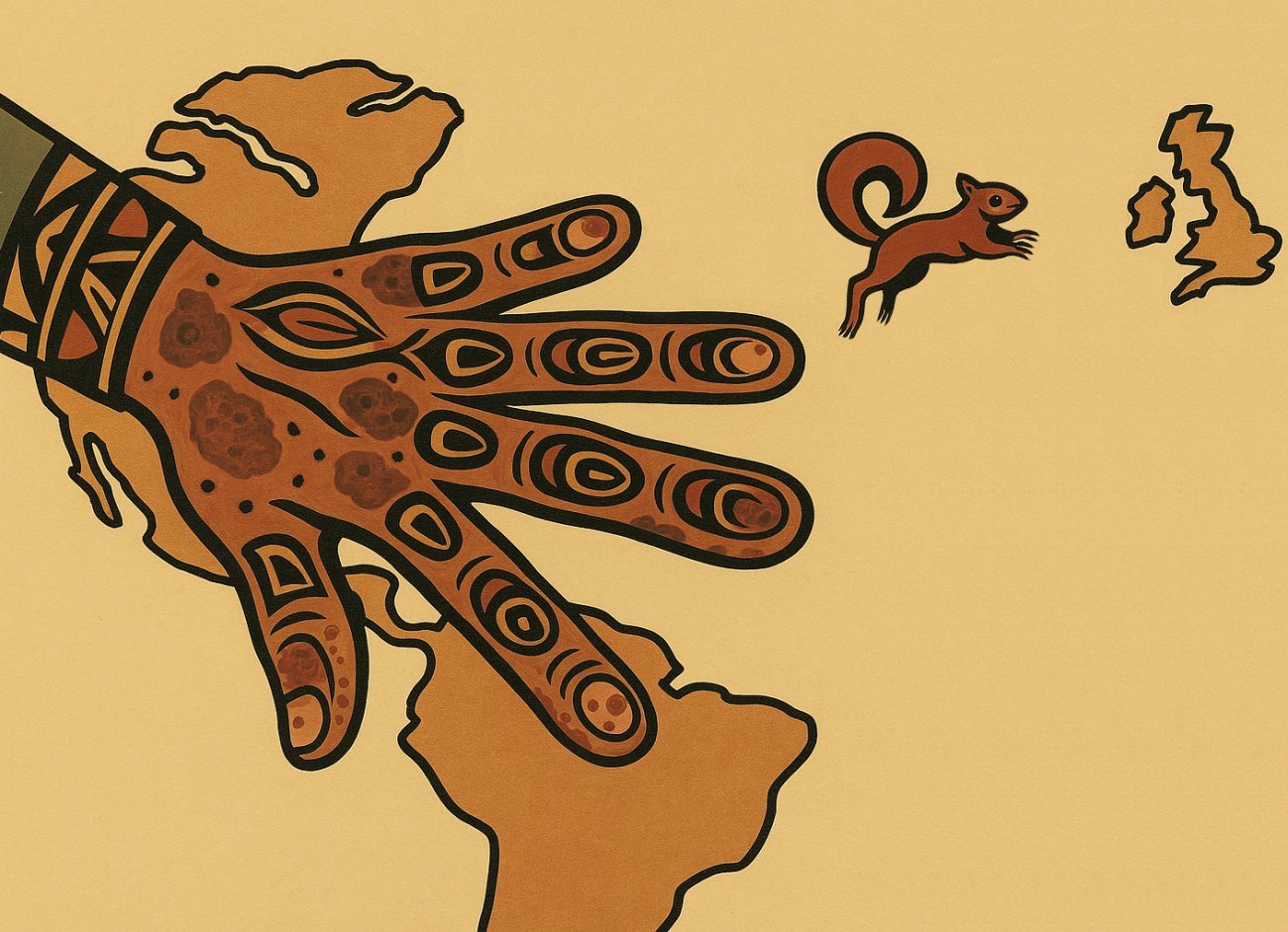
Leprosy existed in America long before the arrival of Europeans: a new study reveals the history of a neglected pathogen.
Long considered a disease brought to the Americas by European colonizers, leprosy may actually have a much older history on the American continent. Scientists from the Institut Pasteur, the CNRS, and the University of Colorado (USA), in collaboration with various institutions in America and Europe, reveal that a recently identified second species of bacteria responsible for leprosy, Mycobacterium lepromatosis, has been infecting humans in the Americas for at least 1,000 years, several centuries before the Europeans arrived. These findings will be published in the journal Science on May 29, 2025.
Leprosy is a neglected disease, mainly caused by the bacterium Mycobacterium leprae, affecting thousands of people worldwide: approximately 200,000 new cases of leprosy are reported each year. Although M. leprae remains the primary cause, this study focused on another species, Mycobacterium lepromatosis, discovered in the United States in 2008 in a Mexican patient, and later in 2016 in red squirrels in the British Isles. Led by scientists from the Laboratory of Microbial Paleogenomics at the Institut Pasteur, also associated with the CNRS, and the University of Colorado, in collaboration with Indigenous communities and over 40 scientists from international institutions including archaeologists, this study analyzed DNA from nearly 800 samples, including ancient human remains (from archaeological excavations) and recent clinical cases presenting symptoms of leprosy. The results confirm that M. lepromatosis was already widespread in North and South America long before European colonization and provide insights into the current genetic diversity of pathogenic Mycobacteria.
"This discovery transforms our understanding of the history of leprosy in America," said Dr. Maria Lopopolo, the first author of the study and researcher at the Laboratory of Microbial Paleogenomics at the Institut Pasteur. "It shows that a form of the disease was already endemic among Indigenous populations well before the Europeans arrived."
The team used advanced genetic techniques to reconstruct the genomes of M. lepromatosis from ancient individuals found in Canada and Argentina. Despite the geographic distance of several thousand kilometers, these ancient strains dating from similar periods (approximately 1,000 years ago) were found to be surprisingly genetically close. Although they belong to two distinct branches in the evolutionary tree of the genus Mycobacterium, these branches are genetically closer to each other than to any other known branch. This genetic proximity, combined with their geographical distance, necessarily implies a rapid spread of the pathogen across the continent, likely within just a few centuries.
The scientists also identified several new lineages, including an ancestral branch that despite having diverged from the rest of the known species’ diversity over 9,000 years ago, it continues to infect humans today in North America — a discovery suggesting an ancient and long-lasting diversification on the continent, as well as a largely unexplored diversity that likely remains to be found.
Notably, the analyses also suggest that the strains found in red squirrels in the UK in 2016 are part of an American lineage that was introduced to the British Isles in the 19th century, where it subsequently spread. This discovery highlights the recent ability of the pathogen to cross continents, likely through human or commercial exchanges.
"We are just beginning to uncover the diversity and global movements of this recently identified pathogen. The study allows us to hypothesize that there might be unknown animal reservoirs," said Nicolás Rascovan, the lead author of the study and head of the Laboratory of Microbial Paleogenomics at the Institut Pasteur. "This study clearly illustrates how ancient and modern DNA can rewrite the history of a human pathogen and help us better understand the epidemiology of contemporary infectious diseases."
The project was conducted in close collaboration with Indigenous communities, which were involved in decisions regarding the use of ancestral remains and the interpretation of results. Ancient DNA and remaining materials were returned when requested, and the generated data was shared via ethical and adaptable platforms designed to allow data sharing that meets the specific expectations of Indigenous communities.
Illustration evoking the history of leprosy in America © Image generated by ChatGPT (OpenAI)
Source
Uncovering pre-European contact leprosy in the Americas and its enduring persistence, Science, May 29, 2025
Maria Lopopolo1,2†, Charlotte Avanzi3†, Sebastian Duchene4, Pierre Luisi1,5,6,7, Alida de Flamingh8, Gabriel Yaxal Ponce-Soto1, Gaetan Tressieres1,9, Sarah Neumeyer1, Frederic Lemoine1, Elizabeth A. Nelson1,§, Miren Iraeta-Orbegozo10, Jerome S. Cybulski11, Joycelynn Mitchell12, Vilma T. Marks13, Linda B. Adams13$, John Lindo14, Michael DeGiorgio15, Nery Ortiz16, Carlos Wiens16, Juri Hiebert16, Alexandro Bonifaz17, Griselda Montes de Oca17, Vanessa Paredes-Solis18, Carlos Franco-Paredes3,19, Lucio Vera-Cabrera20, Jose G. Pereira Brunelli21, Mary Jackson3, John S. Spencer3, Claudio G. Salgado22, Xiang-Yang Han23, Camron M. Pearce3, Alaine K. Warren3, Patricia S. Rosa24, Amanda J. de Finardi24, Andrea de F. F. Belone24, Cynthia Ferreira25,26, Philip N. Suffys27, Amanda N. Brum Fontes27°, Sidra E. G. Vasconcellos27, Roxane Schaub28,29, Pierre Couppie29,30, Kinan Drak Alsibai31, Rigoberto Hernandez-Castro32, Mayra Silva Miranda33, Iris Estrada-Garcia34, Fermin Jurado-Santacruz35, Ludovic Orlando9, Hannes Schroeder10, Lluis Quintana-Murci36,37, Mariano Del Papa38, Ramanuj Lahiri13, Ripan S. Malhi8, Simon Rasmussen39, Nicolas Rascovan1*
1 Institut Pasteur, Université Paris Cité, CNRS UMR 2000, Microbial Paleogenomics Unit, Paris, France
2 Universite de Paris, INSERM, System Engineering and Evolution Dynamics, Paris, France.
3 Mycobacteria Research Laboratories, Colorado State University; Department of Microbiology, Immunology and Pathology, Fort Collins, Colorado, USA
4 Institut Pasteur, Universite de Paris, Evolutionary dynamics of infectious diseases Unit, Paris, France
5 Instituto de Antropologia de Cordoba (IDACOR), CONICET, Universidad Nacional de Cordoba, 5000 Cordoba, Argentina.
6 Departamento de Antropologia, Facultad de Filosofia y Humanidades, Universidad Nacional de Cordoba, 5000 Cordoba, Argentina.
7 Programa de Referencia y Biobanco Genomico de la Poblacion Argentina (PoblAr), Buenos Aires C1425FQD, Argentina
8 Center for Indigenous Science, Carl R. Woese Institute for Genomic Biology, University of Illinois, Urbana-Champaign, Illinois, USA
9 Centre for Anthropobiology and Genomics of Toulouse, CNRS UMR5288, Universite Paul Sabatier, Toulouse, France
10 The Globe Institute, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark
11 Canadian Museum of History, Gatineau, Quebec, Canada
12 Metlakatla First Nation, Prince Rupert, British Columbia, Canada
13 United States Department of Health and Human Services, H 38 ealth Resources and Services Administration, Health Systems Bureau, National Hansen's Disease Program, Baton Rouge, LA, USA
14 Department of Anthropology, Emory University, Atlanta, Georgia, USA
15 Department of Electrical Engineering and Computer Science, Florida Atlantic University, Boca Raton, Florida, USA.
16 Mennonita K81 Hospital D.d.C. 166, Asuncion, Paraguay
17 Servicio de Dermatologia, Hospital General de Mexico "Dr. Eduardo Liceaga", Cd. Mexico, 06720, Mexico
18 Hospital General de Morelia Dr. Miguel Silva, Morelia, Mexico
19 Hospital Infantil de Mexico, Federico Gomez, Cd. Mexico, 06720, Mexico
20 Laboratorio Interdisciplinario de Investigacion Dermatologica, Servicio de Dermatologia, Hospital Universitario “Dr. Jose Eleuterio Gonzalez”, Monterrey, Nuevo Leon, Mexico
21 Leprosy National Program, Ministry of Public Health and Social Welfare, Paraguay
22 Laboratorio de Dermato-Imunologia, Universidade Federal do Para, Marituba, Para, Brazil
23 Department of Laboratory Medicine, Division of Pathology/Lab Medicine, The University of Texas MD Anderson Cancer Center, Houston, TX
24 Research Division, Instituto Lauro de Souza Lima, Bauru, Sao Paulo, Brazil
25 Laboratorio de Biologia Molecular, Fundacao Hospitalar Alfredo da Matta, Manaus, Am, Brazil
26 Programa de Pos-Graduacao em Medicina Tropical, Universidade do Estado do Amazonas, Manaus, Amazonas, Brazil
27 Instituto Oswaldo Cruz, Fundacao Oswaldo Cruz, Rio de Janeiro, Brazil
28 CIC Inserm 1424, Institut sante des populations en Amazonie, Centre Hospitalier de Cayenne, 97300 Cayenne, French Guiana
29 UMR Tropical Biome and Immuno-pathophysiology, Universite de Guyane, Cayenne, French Guiana
30 Department of Dermatology, Cayenne Hospital Centre, Cayenne, French Guiana
31 Department of Pathology and Centre of Biological Resources (CRB Amazonie), Cayenne Hospital Centre, Cayenne, French Guiana
32 Department of Ecology of Pathogens. General Hospital Dr. Manuel Gea Gonzalez, Tlalpan 14080, Mexico City, Mexico
33 CONAHCYT-Instituto Nacional de Enfermedades Respiratorias "Ismael Cosio Villegas". CDMX, Mexico
34 Depto. Inmunologia, Escuela Nacional de Ciencias Biologicas, IPN, CDMX, Mexico
35 Centro Dermatologico "Dr. Ladislao de la Pascua," Secretaria de Salud, Mexico City, Mexico
36 Institut Pasteur, Universite Paris Cite, CNRS UMR2000, Human Evolutionary Genetics Unit, Paris, France
37 Chair Human Genomics and Evolution, College de France, Paris, France
38 Division Antropologia Museo de Ciencias Naturales, FCNyM-UNLP, La Plata, Argentina
39 Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen N, Denmark
°Current address: Department of Science and Technology (Decit), Secretariat of Science, Technology and Innovation and the Economic-Industrial Complex of Health (SECTICS), Brazilian Ministry of Health (MS), Brasilia, Brazil
§Current address: Department of Anthropology, Southern Methodist University, Dallas, Texas, USA
$ retired
†These authors contributed equally to this work.
*Corresponding author.