RNA viruses, like influenza viruses, have broad genetic variability, which gives them a high adaptive potential. As part of the fight against RNA viruses, scientists at the Institut Pasteur – evolutionary virologists – decided to alter their future. Along with colleagues from Lund University (Sweden), they succeeded in altering the genomes of an enterovirus and an influenza virus, thereby reducing their ability to reproduce and be virulent. This method of vaccine design is innovative, targeted and highly precise.
"RNA viruses evolve and adapt easily, due to their broad genetic variability." That is the basis of the study carried out by the Viral Populations and Pathogenesis Unit led by Marco Vignuzzi. "This is why RNA viruses pose a serious threat to human health" explains Gonzalo Moratorio, postdoctoral fellow within the Unit, and first author of the article published in the journal Nature Microbiology, who is soon to return to his his native Uruguay for a new post as Assistant Professor at the Faculty of Science.
And what if the potential to adapt became their weak spot? "We are evolutionary virologists and, as such, we were seeking a means of attenuating these viruses and reducing their chances of success by genetically altering their futures."
Viral population is linked to pathogenesis
We already know that RNA viruses “live and circulate as a population of diverse genetically linked variants", according an article published in Nature in 2006 by Marco Vignuzzi, revealing the link between viral population diversity and pathogenesis. "Viruses mutate from one generation to the next. They mutate a lot. This explains their huge adaptive potential. RNA viruses are even recognized as being the Earth's fastest-evolving organisms." As well as a high mutation rate, they do not have the capacity to correct errors. "This is the price they pay for their adaptability. Most of their mutations are detrimental. Some others, however, make them particularly virulent", adds Gonzalo Moratorio.
Genetic engineering viral genomes to influence mutations
The scientists wanted to increase the proportion of detrimental mutations. Altering the quality of viral replication reduces viral ability to generate progeny that is viable in the environment. This ultimately blocks viral propagation.
"We altered the genomes of the Coxsackie B3 enterovirus and the influenza A virus, in order to redirect their evolutionary trajectories towards detrimental mutations. And it worked! We changed synonymous codons in the viral genomes, coding for the amino acids serine and leucine. These changes caused the viruses to generate more mutations known as "stop" mutations, both in vitro and in vivo. Stop mutations inhibit the virus’ life cycle. But even though not all viruses generated "stop" mutations, these were frequent enough to mean that, overall, the viral population decreased." The scientists therefore merely had to observe the progressive reduction in viral multiplication, which – as expected – led to a marked decline in virulence.
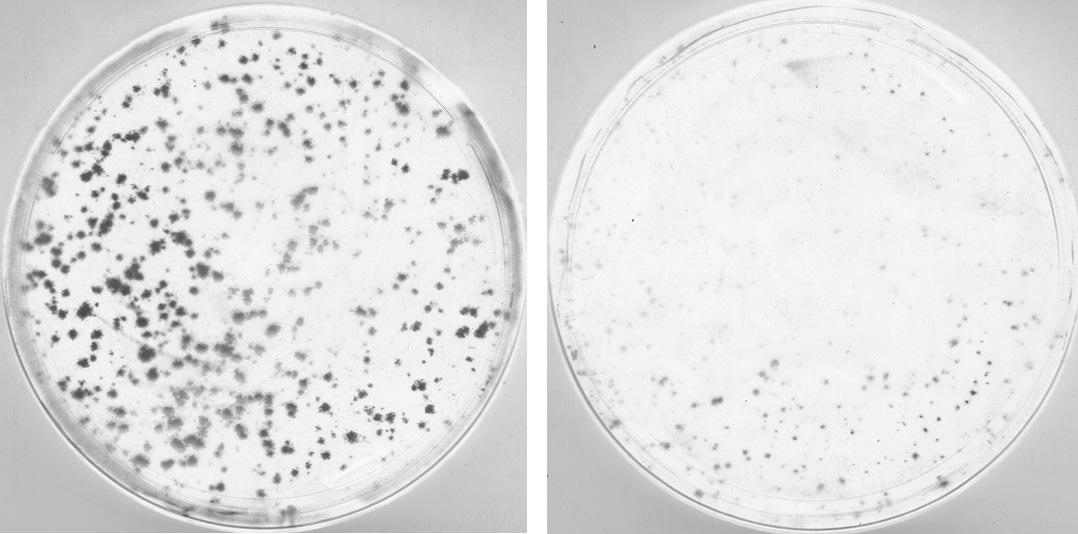
An effective candidate vaccine
This molecular biology technique uses evolutionary concepts to attenuate viruses by genetically altering them, at the same time conferring protection on vaccinated mice. This is an innovative vaccine method, for which the Institut Pasteur has filed a patent application. The scientists proved in vivo that mice vaccinated in this way with candidate vaccines were protected against lethal challenge. "We showed that this strategy may be implemented within the framework of broad spectrum vaccination against RNA viruses." Vaccines remain the most effective way of trying to prevent deaths from RNA viruses, but to date viral vaccines are rare.
This study, which was funded by the Major Federating Program Vaccinology at the Institut Pasteur, a Pasteur-Roux-Cantarini grant and an ERC grant, is the result of mathematicians and biologists working intimately together in the Unit, in a new way they call “computational and mathematical virology”.
Although still at the experimental stage, this research offers major promise in terms of vaccination against RNA viruses. Over the last few years, a number of serious epidemics have been caused by RNA viruses: the chikungunya and Zika viruses in the Americas, coronaviruses in the Middle East and the Ebola virus (report in The Research Journal) in West Africa.
Source
Attenuation of RNA viruses by redirecting their evolution in sequence space, Nature Microbiology, June 5, 2017.
Gonzalo Moratorio1, Rasmus Henningsson1,2,3*, Cyril Barbezange1*, Lucia Carrau1,4, Antonio V. Bordería1,2, Hervé Blanc1, Ste phanie Beaucourt1, EnzoZ.Poirier1,4, Thomas Vallet1, Jeremy Boussier2,5,6, BryanC.Mounce1, Magnus Fontes2,3 and Marco Vignuzzi1
1. Viral Populations and Pathogenesis Unit, Institut Pasteur, CNRS UMR 3569, 28 rue du Dr. Roux, 75724 Paris cedex 15, France.
2. International Group for Data Analysis, Institut Pasteur, C3BI, USR 3756 IP CNRS, 28 rue du Dr. Roux, 75724 Paris cedex 15, France.
3. Centre for Mathematical Sciences, Lund University, 22100 Lund, Sweden.
4. Sorbonne Paris Cité, Université Paris Diderot, Cellule Pasteur, 75013 Paris, France.
5. Unité d’Immunobiologie des Cellules Dendritiques, Institut Pasteur, Inserm 1223, 25 rue du Dr. Roux, 75724 Paris cedex 15, Paris, France.
6. Ecole doctorale Frontières du vivant, Université Paris Diderot, 75013 Paris, France.
*These authors contributed equally to this work.