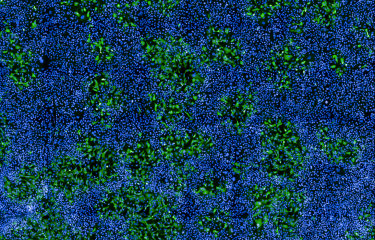
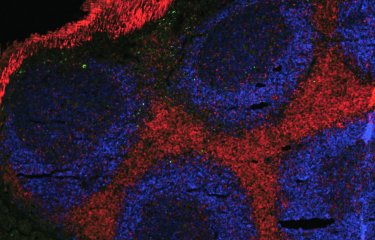
An immune receptor normally known for identifying viral RNA is also capable of binding to cellular RNA to induce immune defenses. This mechanism could trigger immunity against viruses before they become detectable.
RNA viruses, which include influenza viruses, SARS-CoV-2 (which causes COVID-19) and HIV, are one of the major threats that the immune system has to deal with. In a recent study published in the journal iScience, French and US research teams investigated the innate immune mechanisms used by the body in response to RNA viruses. This publication shows that some immune receptors that detect the presence of viral RNA are also activated by RNA produced by our cells, potentially enabling cells to react more quickly to infection.
A conserved evolutionary mechanism against RNA viruses
During an infection, viral RNA is introduced into the cell cytoplasm so that it can be read and translated into proteins needed for viral replication. To counter this replication, human cells express cytoplasmic RIG-I-like receptors, which serve as the first line of defense against viral infections. The RNA molecules bind to these receptors, triggering defense mechanisms in the innate immune system to tackle the infection. It was previously believed that these receptors were only sensitive to viral RNA, but this new study shows that cells are capable of producing endogenous RNA molecules that are also able to activate the receptors. The structure of this endogenous RNA mimics viral RNA so that it can be recognized by RIG-I receptors.
In this study, the scientists studied the immune responses induced by infection with the dengue and measles viruses, which cause acute infections, and also the HIV virus, which causes chronic infection. They showed that the same type of endogenous RNA is involved in triggering innate immunity for different infections, which indicates that this mechanism has been highly conserved during evolution to protect against viral infections. The scientists hypothesize that by recognizing endogenous RNA, the cell is able to keep one step ahead of the infection and induce immunity before the virus has replicated enough to be detected directly. Cells therefore have defense mechanisms that they trigger themselves to ward off infection.
A signaling pathway modulated by viral presence
The results of the study also show that endogenous RNAs are always present in cells but that they do not normally bind to RIG-I receptors and are only detected after the onset of a viral infection. But how do these RNAs manage to signal infection so quickly if the virus has not been detected by receptors? The detailed mechanisms have not yet been elucidated, but it seems that the presence of the virus interferes in certain cellular biochemical pathways that determine the immunogenicity of endogenous RNAs, in other words their ability to trigger an immune response. When the cell is infected, it modifies processes that lead to the expression of endogenous RNAs, making them capable of binding to RIG-I receptors. It is therefore an active process in which the host reacts to viral presence in its cells and uses endogenous RNA to induce an immune response.
This research offers new keys to understanding RNA virus infections and the innate immune response to them. The study particularly describes the case of HIV infection, which takes advantage of this mechanism. The study was based on an international French-American collaboration that combined molecular biology, cell biology and a clinical study of HIV patients to rethink the widely accepted model of immunity.
This study is part of the priority scientific area Emerging infectious diseases of the Institut Pasteur's strategic plan for 2019-2023.
Source:
Y RNAs are conserved endogenous RIG-I ligands across RNA virus infection and are targeted by HIV-1, iScience, July 15, 2022
Nicolas Vabret1,2,10, Valérie Najburg3, Alexander Solovyov12, Ramya Gopal1,2,10, Christopher McClain1,2,10, Petr Šulc4, Sreekumar Balan1,2,10, Yannis Rahou5, Guillaume Beauclair3, Maxime Chazal3, Hugo Varet6,7, Rachel Legendre6,7, Odile Sismeiro6, Raul Y. Sanchez David3, Lise Chauveau8, Nolwenn Jouvenet3, Martin Markowitz9, Sylvie van der Werf5, Olivier Schwartz8, Frédéric Tangy3, Nina Bhardwaj1,2,10,11, Benjamin D. Greenbaum12,13, et Anastassia V. Komarova3,5
1 - Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA
2 - Precision Immunology Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA
3 - Viral Genomics and Vaccination Unit, Department of Virology, Institut Pasteur, Université de Paris, CNRS UMR-3569, 75015 Paris, France
4 - Center for Molecular Design and Biomimetics at the Biodesign Institute and School of Molecular Sciences, Arizona State University, Tempe, AZ 85287, USA
5 - Molecular Genetics of RNA Viruses, Department of Virology, Institut Pasteur, Université de Paris, CNRS UMR-3569, 75015 Paris, France
6 - Transcriptome and EpiGenome Platform, BioMics, Center of Innovation and Technological Research, Institut Pasteur, Université de Paris, 28 rue du Docteur Roux, 75724 Paris Cedex 15, France
7 - Hub Informatique et Biostatistique, Centre de Bioinformatique, Biostatistique et Biologie Intégrative (C3BI, USR 3756 IP-CNRS), Institut Pasteur, Université de Paris, 28 Rue du Docteur Roux, 75724 Paris Cedex 15, France
8 - Virus & Immunity Unit, Department of Virology, Institut Pasteur, Université de Paris, CNRS UMR-3569, 75015 Paris, France
9 - Aaron Diamond AIDS Research Center, TheRockefeller University, New York, NY, USA
10 - Department of Medicine, Hematology and Medical Oncology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA
11 - Extra-mural Member, Parker Institute of Cancer Immunotherapy, USA
12 - Computational Oncology, Department of Epidemiology and Biostatistics, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA
13 - Physiology, Biophysics, & Systems Biology, Weill Cornell Medicine, New York, NY 10065, USA