Press release
Paris, April 14th, 2014
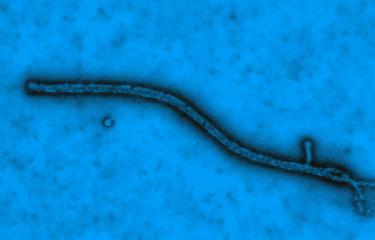
In an article which appeared in The New England journal of Medicine on 16 April, researchers from Inserm (Jean Mérieux-Inserm BSL-4 Laboratory, Lyon) and the Institut Pasteur have published their initial findings on the characteristics of the Ebola virus discovered in Guinea. Initial virological investigations enabled them to identify Zaire ebolavirus as the pathogen responsible for this epidemic. Performed in less than a month, sequencing of the complete genome and subsequent phylogenetic analysis show that the virus present in Guinea forms a clade (variant) that is distinct from strains previously identified in the Democratic Republic of Congo and in Gabon. Epidemiological investigations also linked the laboratory confirmed cases with the initial deaths recorded during the December 2013 outbreak.
Ebola virus is a lethal, highly contagious virus for which there is presently no treatment. The symptoms are somewhat non-specific, and include fever, severe diarrhoea and vomiting. Between 30 and 90% of those infected with this organism die as a result.
On 2 April 2014, the World Health Organisation (WHO), in a communiqué published by the UN, reported that it had recorded 5 new cases of Ebola fever in Guinea. Since January, the total number of suspected and confirmed cases of Ebola fever in the present outbreak in Guinea is 127, with 83 deaths, according to WHO, which states that 35 cases were confirmed by laboratory testing.
The initial samples were analysed in Lyon in the Jean Mérieux-Inserm BSL-4 Laboratory directed by Hervé Raoul, Inserm Research Director, by the French National Reference Centre for Viral Haemorrhagic Fevers (attached to the Biology of Viral Emerging Infections Unit at the Institut Pasteur, directed by Sylvain Baize). A positive diagnosis was made.
A mobile BSL-4 laboratory was deployed in Guinea to provide assistance with diagnosis in the field. This mobile laboratory was developed as part of a European project, “EMLab”, coordinated by German researchers, and in which the Jean Mérieux-Inserm BSL-4 Laboratory is the French partner.
The researchers were able to analyse blood samples from 20 patients. Various tests were conducted by the scientists in order to establish a specific identity card for the virus.
Viral RNA was extracted from the blood samples, and then amplified and sequenced. These sequences were finally compared to 48 already known complete Ebola virus genomes. According to results, the analysis showed 97% identity with strains identified in the Democratic Republic of Congo in 1976 and 2007, and in Gabon in 1994 and 1996.
“These results demonstrate that we are facing the emergence of a new “form” of this virus in Guinea,” explains Hervé Raoul, Director of the BSL-4 Laboratory. This form is common to cases discovered since the month of December.
It would appear that the epidemic originated from a single introduction from animal to human.
Apart from the present epidemic, these results show that the endemic area for Ebola virus is greater than previously known, and that as a consequence, West Africa should henceforth be considered an area of risk for Ebola virus. Measures aimed at preventing transmission from wild fauna to humans and quickly identifying such events if there is a recurrence must be put in place in Guinea, as well as in neighbouring countries.
Further information on the BSL-4 laboratory
The Jean Mérieux BSL-4 Laboratory is a high-level containment laboratory dedicated to the study of Class 4 pathogens. The biological safety level applied is 4, the highest possible level. The researchers working there wear a full body, air-supplied, positive pressure suit to protect them from all contamination. The laboratory is itself maintained under negative pressure in order to protect the environment. Moreover, all wastes produced are completely inactivated, and the exhaust air is purified by a double absolute filtration system. This laboratory currently remains the structure offering the largest experimental capacity in Europe for this containment level.
Highly pathogenic agents:
Class 4 pathogens (or risk group 4) are highly pathogenic microorganisms characterised by a very high mortality rate, a lack of prophylactic or therapeutic measures to provide protection, and ready transmissibility. All class 4 pathogens currently known are viruses, and include viruses that cause haemorrhagic fevers or encephalitis. They include the Ebola, Marburg, Lassa, Junin, Machupo, Guanarito, Sabia, Crimean-Congo, Nipah and Hendra viruses.
To obtain photographs of the BSL-4 laboratory : http://www.serimedis.inserm.fr/fr/feature/content/id/989
© Inserm/Guénet François
Sources
Emergence of Zaire Ebola Virus Disease in Guinea — Preliminary Report
Sylvain Baize, Ph.D., Delphine Pannetier, Ph.D., Lisa Oestereich, M.Sc.,Toni Rieger, Ph.D., Lamine Koivogui, Ph.D., N’Faly Magassouba, Ph.D., Barrè Soropogui, M.Sc., Mamadou Saliou Sow, M.D., Sakoba Keïta, M.D., Hilde De Clerck, M.D., Amanda Tiffany, M.P.H., Gemma Dominguez, B.Sc.,
Mathieu Loua, M.D., Alexis Traoré, M.D., Moussa Kolié, M.D., Emmanuel Roland Malano, M.D., Emmanuel Heleze, M.D., Anne Bocquin, M.Sc., Stephane Mély, M.Sc., Hervé Raoul, Ph.D., Valérie Caro, Ph.D., Dániel Cadar, D.V.M., Ph.D., Martin Gabriel, M.D., Meike Pahlmann, Ph.D., Dennis Tappe, M.D., Jonas Schmidt-Chanasit, M.D., Benido Impouma, M.D., Abdoul Karim Diallo, M.D., Pierre Formenty, D.V.M., M.P.H., Michel Van Herp, M.D., M.P.H., and Stephan Günther, M.D.
New England Journal of Medicine, 16 avril 2014, http://dx.doi.org/10.1056/NEJMoa1404505