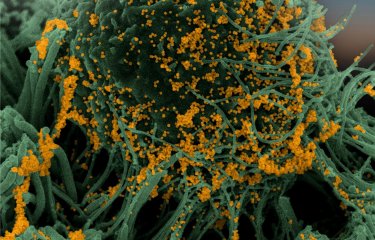
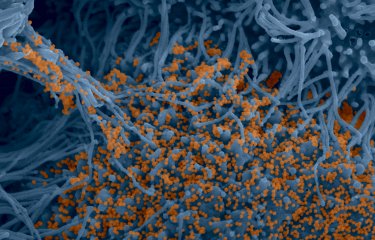
Despite concerted planetary efforts to sequence and analyze SARS-CoV-2, our understanding of the virus is limited by a lack of genomic data on coronaviruses. A group of scientists who began working together at a hackathon have discovered a number of novel coronaviruses by analyzing all the RNA sequencing data that is publicly available at global level.
Current biotechnological tools have been used to sequence the genomes of numerous viruses and their hosts. The sequences produced by laboratories worldwide are freely accessible on the Sequence Read Archive run by the NCBI (National Center for Biotechnology Information) and hosted by the National Institutes of Health (NIH) in the United States.
An infrastructure to make use of available viral sequences
There is an impressive quantity of data available, but these data need to be able to be explored and used. At the hackseqRNA hackathon organized by the University of British Columbia in Vancouver, scientists from a range of different backgrounds met and began working on a project proposed by Artem Babaian, an independent researcher and one of the organizers of the hackathon. The project is a database known as Serratus, which will be made freely available via open access. "Serratus is an open science project for the discovery of new viral sequences via a very large-scale search of all publicly available RNA sequencing, metagenomic, metatranscriptomic and environmental data," explains Rayan Chikhi, Head of the Sequence Bioinformatics laboratory at the Institut Pasteur and co-last author of the study.
To perform this ultra-high-throughput search, the scientists used the Amazon cloud and launched analyses on more than 22,000 processors at the same time. Using innovative methods, they were able to reach a processing rate of more than a million samples per day, for a very low cost of approximately $0.01 per sample.
Novel coronaviruses discovered with Serratus
The algorithms developed enabled the scientists to analyze 5.7 million sequencing samples. They identified thousands of samples containing coronaviruses and discovered several coronavirus species that had not previously been recorded. The analysis also significantly enlarged the number of known RNA viruses.
As well as the source codes, all the raw and processed data generated by Serratus are available free of charge in an open database developed by the team, so that the viral sequences can be analyzed more quickly by other scientists.
"Expanding the known repertoire of coronaviruses and other viruses will enable us to monitor their spread between animals and to humans, thereby helping to avoid further pandemics," concludes Rayan Chikhi.
Source :
Petabase-scale sequence alignment catalyses viral discovery, Nature, January 26, 2022
Robert C. Edgar1 , Jeff Taylor1 , Victor Lin1 , Tomer Altman2 , Pierre Barbera3 , Dmitry Meleshko4,5, Dan Lohr1 , Gherman Novakovsky6 , Benjamin Buchfink7 , Basem Al-Shayeb8 , Jillian F. Banfield9 , Marcos de la Peña10, Anton Korobeynikov4,11, Rayan Chikhi12, and Artem Babaian1,
1 Unaffiliated
2 Altman Analytics LLC, San Francisco, California, USA
3 Computational Molecular Evolution Group, Heidelberg Institute for Theoretical Studies, Heidelberg, Germany
4 Center for Algorithmic Biotechnology, St. Petersburg State University, St. Petersburg, Russia
5 Tri-Institutional PhD Program in Computational Biology and Medicine, Weill Cornell Medical College, New York, USA
6 Department of Medical Genetics, University of British Columbia. Vancouver, BC, Canada
7 Computational Biology Group, Max Planck Institute for Developmental Biology, T¨ubingen, Germany
8 Department of Plant and Microbial Biology, University of California, Berkeley, USA
9 Department of Earth and Planetary Science, University of California, Berkeley, USA
10 Instituto de Biolog´ıa Molecular y Celular de Plantas, Universidad Polit´ecnica de Valencia-CSIC, Valencia, Spain
11 Department of Statistical Modelling, St. Petersburg State University, St. Petersburg, Russia
12 Institut Pasteur, CNRS, Paris, France