In collaboration with New England Biolabs, the DNA Replication research group led by Ludovic Sauguet at the Institut Pasteur has published in Nucleic Acids Research an innovative approach based on PacBio long-read sequencing to precisely characterize the errors made by DNA polymerases with unprecedented accuracy.
DNA polymerases (DNAPs), essential enzymes for DNA replication and repair, are the cornerstone of numerous biotechnological applications, from PCR and sequencing to cloning and gene therapies. Despite their remarkable accuracy, these enzymes occasionally introduce errors with potentially significant consequences, whether evolutionary or pathological, especially in cancer development. Understanding the mechanisms underlying these errors is therefore a major challenge for advancing our knowledge and optimizing their use.
Promising Biotechnological Perspectives
This work opens interesting prospects for the design of tailor-made enzymes, adapted to specific applications requiring either enhanced fidelity or, conversely, controlled mutagenesis, with applications in biotechnology, therapeutics, and diagnostics.
A Novel Methodological Approach
The main innovation of this study lies in the use of a high-throughput method exploiting Pacific Biosciences (PacBio) long-read sequencing without the need for PCR amplification. Unlike previous techniques, this approach is unbiased by the sequence of the template DNA, enabling direct measurement of error rates and detailed mapping of enzyme-specific error profiles. Replicative DNAPs possess dual catalytic activities: a polymerase activity to synthesize DNA and an exonuclease proofreading activity to remove misincorporated nucleotides. The developed method allows integrated evaluation of the efficiency of both mechanisms.
Family-specific Error Profiles Revealed
The researchers analyzed the four main families of replicative DNA polymerases (A, B, C, and D), revealing diverse and family-specific error profiles. This detailed characterization highlights distinct molecular mechanisms governing the fidelity of these enzymes, thereby providing an in-depth understanding of their respective modes of action.
Factors Influencing Enzymatic Fidelity
The study also identified several critical parameters affecting DNAP precision. For example, the ratios of the four deoxyribonucleotide triphosphates (dATP, dGTP, dCTP, dTTP) present in the reaction mix significantly influence the error rate, suggesting that the balance of available substrates directly modulates replication fidelity. The components of the replication system and mutations in the enzyme’s active sites—both in the polymerase domain and the exonuclease domain—were also examined, revealing how structural modifications can impair or enhance enzymatic accuracy.
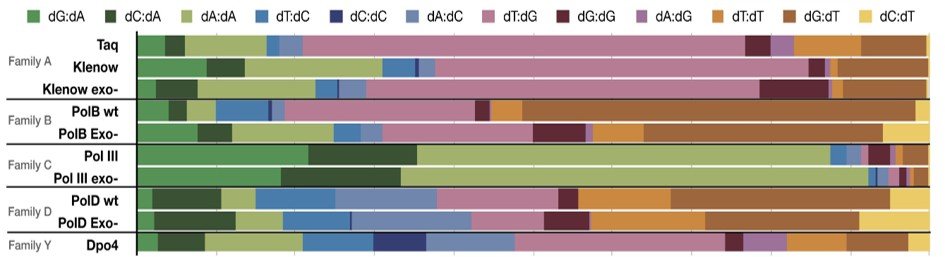
DNA polymerase error rates and profiles. (A) Fidelity measurements in substitutions in a million bases for the different families of DNA polymerases and their respective exo- mutant. (B) Substitution spectra for each DNA polymerase, with dA misincorporations in green, dC misincorporations in blue, dG misincorporations in pink, and dT misincorporations in orange. Legend corresponds to template strand:nascent strand mispair (i.e. dG:dA = template strand dG, nascent strand dA).





