Artemisinin is currently the most effective malarial treatment available. However, the recent emergence in South-East Asia of artemisinin-resistant parasites strengthens the urgent need to identify a new generation of antimalarial drugs. In this context, scientists at the Institut Pasteur and CNRS have determined the three-dimensional structure of a promising new therapeutic target for malaria: SUB1. This protein plays a crucial role in parasite egress from host cells followed by its invasion of new cells. These results, which were published in Nature Communications on September 10, identify the SUB1 activation mechanism - the initial stage that triggers parasite egress from host cells. The structural information provided by the research also furthers the effort to find new antimalarial candidate drugs, as it helps optimize the development of SUB1 inhibitors.
Press release
Paris, September 10, 2014
Nearly 40% of the world's population is exposed to malaria. This disease is caused by parasites of the Plasmodium genus affecting around 220 million people each year and causing nearly 660,000 deaths. Artemisinin has to date been considered the last effective treatment against malaria, and the recent emergence in South-East Asia of artemisinin-resistant parasites represents a real threat to the success of efforts to fight this disease.
After being injected into the host via a bite from an Anopheles mosquito, malaria-causing parasites invade liver cells, the hepatocytes. There the parasites multiply before eventually exiting the hepatocytes in order to infect red blood cells.
Scientists in the team led by Jean-Christophe Barale (Malaria Biology and Genetics Unit, Institut Pasteur and CNRS) have unraveled the spatiotemporal regulation mechanism of the parasite protein known as SUB1, which plays a key part in triggering the egress of parasites from hepatocytes and red blood cells. SUB1 activates various molecules involved in the rupture of the double membrane that surrounds intracellular parasites, thus enabling their release. SUB1 also prepares the newly released parasites to invade new red blood cells. This is how Plasmodium spreads through the bloodstream and thereby brings about the onset of malaria symptoms. The most widely known of these symptoms, high fever, is closely associated with the phase of parasite egress from red blood cells.
SUB1: an innovative therapeutic and preventive target
SUB1 is an innovative therapeutic target, as blocking its action in the hepatic stages would prevent the appearance of blood stage parasites, and therefore the symptoms of malaria. Blocking SUB1 after a blood phase has already started would halt the proliferation of parasite blood stages. A SUB1 inhibitor would therefore have a dual role: a preventive role, via its action during the hepatic stages, and a therapeutic role, via its action during the blood stages of Plasmodium.
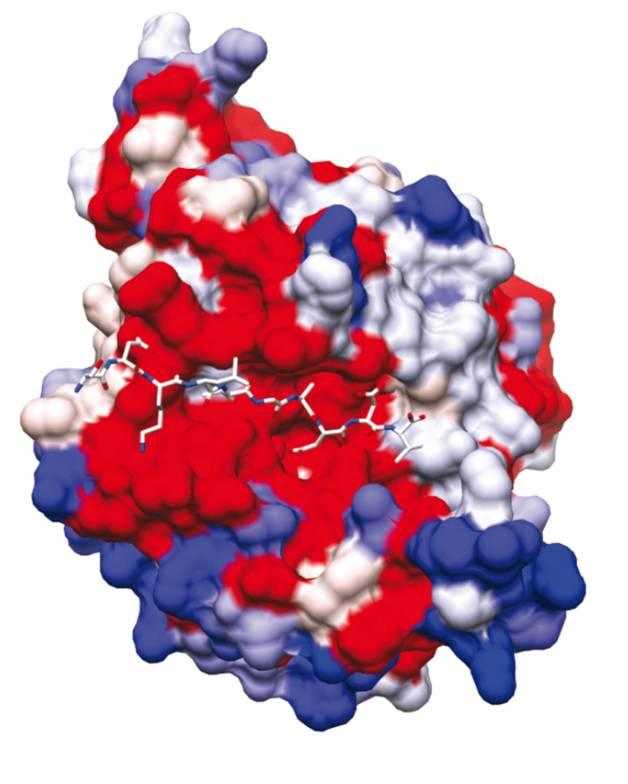
SUB1: identification of a blocking mechanism to prevent parasite egress
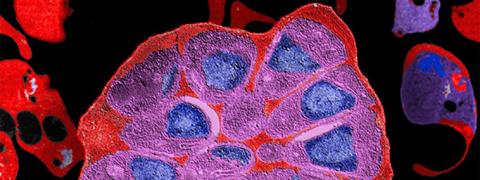
In this study, with the aim of refining research into molecules with the potential to inhibit SUB1 as a trigger for parasite egress, scientists in the teams led by Pedro Alzari and Jean-Christophe Barale (Structural Microbiology Unit and Malaria Biology and Genetics Unit at the Institut Pasteur and CNRS) have succeeded in establishing the three-dimensional structure of SUB1 in its form immediately before parasites are released.
The role of SUB1 is to cut various proteins at very specific sites, thereby activating them and enabling them to rupture the double membrane around the parasite. Rather like a key fitting a lock, these proteins specifically fit the catalytic domain of SUB1, where they are cut and activated.
Scientists at the Institut Pasteur and CNRS have demonstrated that the "lock" - in other words the SUB1 catalytic domain - is naturally protected by a belt, which renders SUB1 inoperative. They have deciphered the sophisticated stratagem that allows SUB1 to self-remove this protective belt, thereby freeing the catalytic domain (the "lock") at the precise moment the parasites are mature and ready to leave the cell in order to invade new cells. They have shown that a significant increase in calcium level within the cell is responsible for freeing and activating the SUB1 "lock", thereby triggering the process of parasite egress. This work has shed light on one of the unlocking mechanisms that trigger the release of mature parasites.
More importantly, in terms of potential applications, these studies provide a highly precise description of the structure and composition of the SUB1 catalytic domain: this gives us a precise idea of the form that chemical compounds should have to access this lock, block the SUB1 "lock" and prevent parasite egress. These findings are the result of a multidisciplinary approach, and represent a crucial stage in research aimed at finding SUB1 inhibitors that could be the basis for a new generation of antimalarial drugs.
Illustrations:
Infected erythrocyte by Plasmodium falciparum parasite, © Institut Pasteur / Biologie des Interactions Hôte-Parasite
The three-dimensional structure of SUB1. Centrally, in red: SUB1 catalytic domain. © Jean-Christophe Barale, Institut Pasteur
This study received funding from the French National Research Agency, the Carnot Pasteur Infectious Diseases Institute, the Fond dédié : Combattre les maladies parasitaires (Sanofi / the French Ministry of Research) and the French Ministry of Defense (DGA).
Source
A novel Plasmodium-specific prodomain fold regulates the malaria drug target SUB1 subtilase, Nature Communications, September 10, 2014. DOI : NCOMMS5833
David Giganti(1*), Anthony Bouillon(2*), Lina Tawk(2), Fabienne Robert(2), Mariano Martinez(1), Elodie Crublet(3), Patrick Weber(3), Christine Girard-Blanc(3), Stéphane Petres(3), Ahmed Haouz(3), Jean-François Hernandez(4), Odile Mercereau-Puijalon(2), Pedro M. Alzari(1,3)& Jean-Christophe Barale(2).
(1) Institut Pasteur, Unité de Microbiologie Structurale, Département de Biologie Structurale et Chimie & CNRS UMR 3528, F-75015 Paris, France
(2) Institut Pasteur, Unité d’Immunologie Moléculaires des Parasites, Département de Parasitologie et de Mycologie & CNRS URA 2581, F-75015 Paris, France
(3) Institut Pasteur, Proteopole & CNRS UMR 3528, F-75015 Paris, France
(4) Institut des Biomolécules Max Mousseron, UMR5247, CNRS, Universités Montpellier 1 & 2, Faculté de Pharmacie 15 avenue Charles Flahault, 34093 Montpellier cedex 5, France
*These authors contributed equally to this work.