In association with CEA-Genoscope and the Sanger Institute, scientists at the Institut Pasteur in Paris, the CNRS, INSERM, the Institut Pasteur of Lille, and Université Lille 2 have recently determined the origin of the emergence of the Mycobacterium tuberculosis bacterium, the main causative agent of tuberculosis. Researchers have also provided insights into its evolutionary success. They have identified several genetic mechanisms that could have contributed to the worldwide dissemination of this pathogen, which currently infects up to 2 billion people. This research, published on the Nature Genetics website on January 6, offers possibilities for identifying new targets in the fight against tuberculosis.
Press release
Paris, january 7, 2013
In collaboration with CEA-Genoscope and the Sanger Institute, the teams working under Roland Brosch , at the Institut Pasteur in Paris, and Philip Supply , at the Institut Pasteur of Lille, have successfully traced the evolutionary history of M. tuberculosis. Their study has confirmed the theory formed during previous research, which suggests that M. tuberculosis strains are genetically highly conserved and have their origins in the same evolutionary branch as Mycobacterium canettii (M. canettii). Mycobacterium canettii strains, which also cause tuberculosis, present a wide genetic diversity and certain genomic traits that point to much older origins. Scientists are also offering possible insights into the factors that have contributed to the evolutionary success of M. tuberculosis and turned tuberculosis into a global pandemic, whereas the strains of M. canettii have remained largely contained within Eastern Africa.
The team of researchers, led by Philip Supply and Roland Brosch, suggests in particular that the strains of M. tuberculosis acquired their virulence and persistence via a combination of several genetic mechanisms, such as loss of function of certain genes, or the acquisition of new genes via horizontal transfer. Having identified genes potentially involved in M. tuberculosis, this work opens new possibilities in the fight against a disease that has become a WHO priority.
--
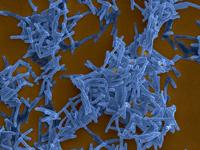
Illustration - Copyright Institut Pasteur
Caption: Mycobacterium tuberculosis, causative agent of tuberculosis. Scanning electron microscopy
Source
Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis, Nature Genetics, January 7, 2013.
Philip Supply 1,2,3,4, Michael Marceau 1,2,3,4, Sophie Mangenot 5, David Roche 5,6, Carine Rouanet 1,2,3,4, Varun Khanna 7, Laleh Majlessi 8,9, Alexis Criscuolo 10, Julien Tap 10, Alexandre Pawlik 7, Laurence Fiette 11,12, Mickael Orgeur 7, Michel Fabre 13, Cécile Parmentier 7, Wafa Frigui 7, Roxane Simeone 7, Eva C. Boritsch 7, Anne-Sophie Debrie 1,2,3,4, Eve Willery 1,2,3,4, Danielle Walker 14, Michael A. Quail 14, Laurence Ma 15, Christiane Bouchier 15, Grégory Salvignol 5,6, Fadel Sayes 8,9, Alessandro Cascioferro 7, Torsten Seemann 16, Valérie Barbe 5, Camille Locht 1,2,3,4, Maria-Cristina Gutierrez 1,2,3,4,17, Claude Leclerc 8,9, Stephen D. Bentley 14, Timothy P. Stinear 18, Sylvain Brisse 10, Claudine Médigue 5,6, Julian Parkhill 14, Stéphane Cruveiller 5,6 & Roland Brosch 7.
1 Institut National de la Santé et de la Recherche Médicale (INSERM), U1019, Center for Infection and Immunity of Lille, Lille, France;
2 Centre National de la Recherche Scientifique (CNRS), Unite mixte de recherche (UMR) 8204, Center for Infection and Immunity of Lille, Lille, France;
3 Univ Lille Nord de France, Université Lille 2, Center for Infection and Immunity of Lille, Lille, France;
4 Institut Pasteur de Lille, Center for Infection and Immunity of Lille, Lille, France;
5 Commissariat à l'Energie Atomique et aux Energies Alternatives CEA/DSV/IG/Genoscope, LABGeM, Evry, France;
6 CNRS-UMR 8030, Evry, France;
7 Institut Pasteur, Unit for Integrated Mycobacterial Pathogenomics, Paris, France;
8 Institut Pasteur, Unité de Régulation Immunitaire et Vaccinologie, Paris, France;
9 INSERM U1041, Paris, France;
10 Institut Pasteur, Genotyping of Pathogens and Public Health (PF8), Paris, France;
11 Institut Pasteur, Unité d'Histopathologie Humaine et Modèles Animaux, Paris, France;
12 Université Versailles-Saint Quentin en Yvelines, Faculté de Médecine, DER Histologie, Versailles, France;
13 Laboratoire de Biologie Clinique, HIA Percy, Clamart, France;
14 Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK;
15 Institut Pasteur, Genopole, Platform Genomics PF1, Paris, France;
16 Victorian Bioinformatics Consortium, Monash University, Clayton, Australia;
17 Institut Pasteur, Département Infection et Epidémiologie, Paris, France;
18 Department of Microbiology and Immunology, University of Melbourne, Parkville, Australia.
Contacts
Institut Pasteur Press Office
Nadine Peyrolo - +33 (0)1 45 68 81 46
Jérémy Lescene - +33 (0)1 45 68 81 01
presse@pasteur.fr
Institut Pasteur Lille Communications Department
Marie-José Hermant - + 33 (0)3 20 87 78 08
marie-jose.hermant@pasteur-lille.fr
Eugénie Devendeville - + 33 (0)3 20 87 77 38
eugenie.dumeignil@pasteur-lille.fr



