A collaborative project conducted by scientists from the Institut Pasteur in Cambodia, the Institut Pasteur in Paris, the Institut Cochin, the Laboratory of Applied Mathematics at Paris Descartes University, Columbia University (New York, USA) and the National Malaria Control Program in Cambodia has identified a molecular marker able to detect Plasmodium falciparum malaria parasites resistant to piperaquine, a drug currently used in combination with artemisinin derivatives in the latest generation of antimalarial treatments recommended by the WHO. This discovery should pave the way for rapid monitoring strategies and enable public health policy makers to recommend effective antimalarial treatments adapted to the local epidemiological situation. This research will be published on November 3, 2016 in the journal The Lancet Infectious Diseases.
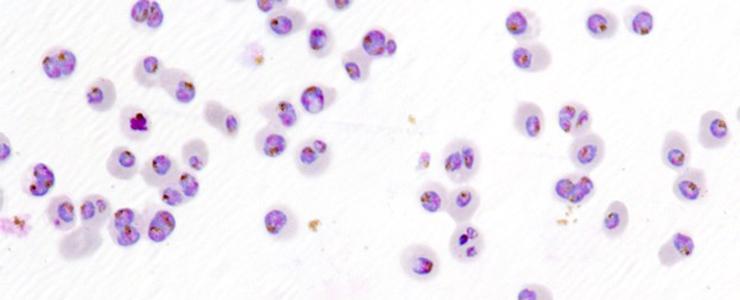
Parasites (violet and blue) in red blood cells (light pink) © Institut Pasteur
Almost 3.2 billion people – virtually half the world's population – are at risk of contracting malaria, a disease caused by Plasmodium parasites for which there is no vaccine yet. There are more than 200 million malaria cases each year and nearly 438,000 deaths. Over the last decade, parasites resistant to artemisinin derivatives used in combination with a partner drug in Artemisinin-based Combination Therapies (ACTs) have emerged in the Mekong basin, endangering the efforts to fight the disease. One of the main threats is that these resistant parasites spread to Sub-Saharan Africa, the region most affected by malaria, as has happened in the past with parasites resistant to treatments based on chloroquine or antifolates.
Plasmodium falciparum malaria episodes are currently treated using ACTs that combine an artemisinin derivative with a long half-life drug (amodiaquine, sulfadoxine/pyrimethamine, mefloquine, lumefantrine or piperaquine). Since 2008, resistance to artemisinin derivatives has been observed in Southeast Asia. This partial resistance does not affect the efficacy of ACTs as long as the partner drugs remain fully effective, and most patients treated by ACTs are cured. However, in some regions of Cambodia and Thailand, multidrug resistant Plasmodium falciparum, surviving both the artemisinin and the partner drugs has recently emerged. This has led to an alarming situation, with increasing difficulties in treating Plasmodium falciparum malaria in these two countries. The spread of these multi-drug resistant strains to other regions would be a public health catastrophe with disastrous consequences.
Cambodia is facing high clinical failure rates (up to 60% in some regions) in patients treated with dihydroartemisinin-piperaquine, since P. falciparum parasites are now able to resist both drugs. Scientists from the Institut Pasteur have recently demonstrated that resistance to artemisinin was strongly associated with mutations in the parasite's K13 gene, but no molecular marker had been identified yet to detect resistance to piperaquine.
Following on from this research, the same Institut Pasteur teams from Cambodia and Paris, working in the new "Malaria Translational Research international joint unit", in collaboration with scientists from the Institut Cochin, the Laboratory of Applied Mathematics (MAP5) at Paris Descartes University, Columbia University (New York, USA) and the National Malaria Control program in Cambodia with the support of the WHO, have identified a molecular marker that is strongly associated with Plasmodium falciparum resistance to piperaquine. This molecular "signature" is characterized by an increase in the copy number (amplification) of two genes, plasmepsin 2 and 3, encoding enzymes involved in hemoglobin degradation. The article released in the The Lancet Infectious Diseases journal reports that individuals infected with parasites with multiple copies of the plasmepsin 2 gene and any mutation in the K13 gene, (which confers resistance to artemisinin) are on average 20 times more likely to experience dihydroartemisinin-piperaquine treatment failure. This work also shows that the amplification of these genes systematically coincides with a reduction in the number of copies of the mdr1 (multi-drug resistance-1) gene, whose amplification is associated with resistance to mefloquine. The authors therefore suspect that the emergence of piperaquine resistance coincides with regained efficacy of mefloquine.
This finding should pave the way for rapid monitoring strategies that enable public health policy makers to recommend effective antimalarial treatments adapted to the epidemiological situation. In addition to the K13 marker, the identification of the piperaquine resistance marker should allow scientists to provide a global map of multi-drug resistant parasites, define high resistance areas and implement effective treatments and appropriate control measures.
This research was supported by the Institut Pasteur in Cambodia, the Institut Pasteur in Paris, the French National Research Agency, the "Investing in the Future" program, the "Integrative Biology of Emerging Infectious Diseases" Laboratory of Excellence (grant 460 no. ANR-10-LABX-62-IBEID), the National Institutes of Health (R01 AI109023 and AI124678) and WHO (KARMA 2 project).
Source
A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria, The Lancet Infectious Diseases, November 3, 2016
Benoit Witkowski*1,2, Valentine Duru*1, Nimol Khim1,2, Leila S. Ross3, Benjamin Saintpierre4, Johann Beghain4, Sophy Chy1, Saorin Kim1, Sopheakvatey Ke1, Nimol Kloeung1, Rotha Eam1, Chanra Khean1, Malen Ken1, Kaknika Loch1, Anthony Bouillon2,5, Anais Domergue1, Laurence Ma6, Christiane Bouchier6, Rithea Leang7, Rekol Huy7, Grégory Nue8l, Jean-Christophe Barale2,5, Eric Legrand4, Pascal Ringwald9, David A. Fidock3, Odile Mercereau-Puijalon4, Frédéric Ariey10, Didier Ménard1,2.
1 Malaria Molecular Epidemiology Unit, Institut Pasteur in Cambodia, Phnom Penh, Cambodia
2 Malaria Translational Research Unit, Institut Pasteur, Paris, France - Institut Pasteur in Cambodia, Phnom Penh, Cambodia
3 Department of Microbiology and Immunology and Division of Infectious Diseases, Department of Medicine, Columbia University Medical Center, New York, USA
4 Department of Parasites and Insect Vectors, Institut Pasteur, Paris, France
5 Structural Microbiology Unit, Biology of Malaria Targets Group, Department of Structural Biology and Chemistry and CNRS, UMR3528, Institut Pasteur, Paris, France.
6 Plate-forme Génomique, Département Génomes et Génétique, Institut Pasteur, Paris, France
7 National Center for Parasitology, Entomology and Malaria Control, Phnom Penh, Cambodia
8 Laboratoire de Mathématiques Appliquées (MAP5) UMR CNRS 8145, Université Paris Descartes, Paris, France
9 Global Malaria Programme, World Health Organization, Geneva, Switzerland.
10 Institut Cochin Inserm U1016, Université Paris-Descartes, Sorbonne Paris Cité, and Laboratoire 54 de Parasitologie-Mycologie, Hôpital Cochin, Paris, France