Gene structure, including the transcript leader, is very diverse in fungi. New sequencing data analysis from the RNA Biology of Fungal Pathogens unit revealed that in the pathogenic yeasts Cryptococcus, the transcript leader sequence are rich in potential upstream Open Reading Frames, which regulate both gene expression and protein diversity.
Pathogenic Cryptococcus species are responsible for nearly 200 000 deaths every year in the word. More knowledge of the gene structure is still needed to understand their regulation and their individual impact on virulence. In this study, scientists from the unit RNA Biology of Fungal Pathogens used a large set of different types of RNA sequence data to re-annotate precisely the extremities of each coding genes in two pathogenic fungi (i.e Cryptococcus neoformans and C. deneoformans). Surprisingly, the analysis of the structure of the Cryptococcus transcript leader sequences revealed the presence of more than 10 000 potential upstream Open Reading Frame (uORF). We showed here that in these fungi they are a major contributor to translation repression.
Eukaryotic protein synthesis initiates at a start codon defined by an AUG and its surrounding Kozak sequence context, but studies of S. cerevisiae suggest this context is of little importance in fungi. Nevertheless, in Cryptococcus uORF use depends on the Kozak sequence context of its start codon, and uORFs with strong contexts promote nonsense-mediated mRNA decay. Alternative usage of translation start site is also predicted to be a means to diversify the proteomes in these yeasts. Thus, numerous Cryptococcus mRNAs encode predicted dual-localized proteins, including many aminoacyl-tRNA synthetases, in which a leaky AUG start codon is followed by a strong Kozak context in-frame AUG, separated by mitochondrial-targeting sequence (Figure). In contrast, in Saccharomyces cerevisiae, the transcript leader sequences are short and less than one thousand uORF are present. Analysis of other fungal species shows that such dual-localization is also predicted to be common in the diverged mould, Neurospora crassa. Kozak-controlled regulation is correlated with insertions in translational initiation factors in fidelity-determining regions that contact the initiator tRNA. Thus, start codon context is a signal that programs the expression and structures of proteins in fungi.
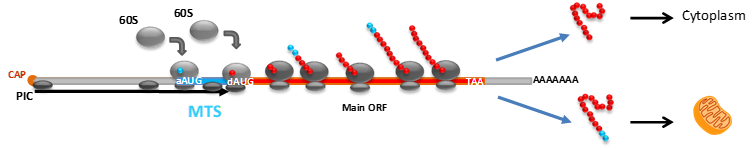
Alternative usage of start codon depending on the sequence and position context can regulate protein targeting in fungi.


