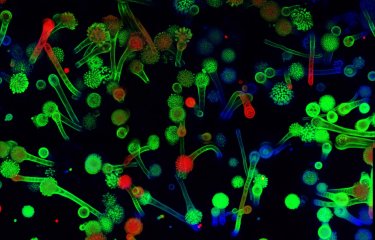
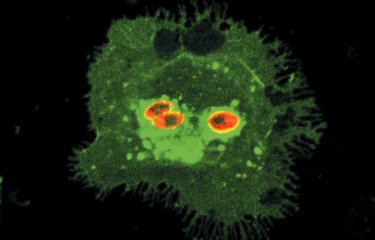
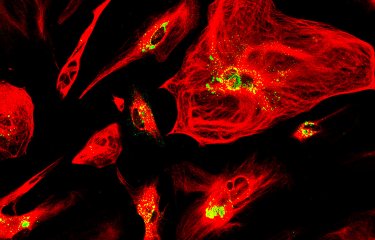
A team of scientists has revealed a gene (CSA6) that regulates genome stability in Candida albicans and other related fungi. The protein produced by this gene, Csa6, therefore represents a potential target for antifungal therapy, at a time when there is an urgent need for the development of novel treatments for fungal infections.
Candida albicans is a fungus found in most healthy individuals; it generally colonizes the mucosa in the digestive and genital tracts. But in some immunocompromised individuals, such as people with HIV or cancer, it becomes pathogenic. It eludes the host's defenses and even causes systemic infections that can be fatal (see the fact sheet on Candidiases). These infections are difficult to treat because there are few classes of antifungal drugs. To make things worse, cases of resistance of C. albicans (and related species) to these drugs are emerging. The resistance of C. albicans to antifungals is exacerbated by the high degree of plasticity of its genome, in other words its ability to modify the number of copies of its chromosomes and the way in which they are organized.
CSA6 gene, a critical cell cycle regulator
In a recent collaborative study, scientists from the Institut Pasteur in Paris and the Jawaharlal Nehru Centre for Advanced Scientific Research (JNCASR) in India examined the effect of overexpression of more than a thousand C. albicans genes, taken individually, on the stability of its genome. This approach enabled them to identify a critical cell cycle regulator.
"Of six genes identified as being involved in genome plasticity, CSA6 caught our attention. Its function was not yet known, but the modulation of its level of expression had an impact on cell survival by negatively affecting cell cycle function. If inactivated, C. albicans stops growing, but if overexpressed, it results in changes to cell morphology, inducing polarized cell growth," explains Christophe d'Enfert, joint last author of the article and Head of the Fungal Biology and Pathogenicity Unit at the Institut Pasteur. This unit is associated with the Institut Pasteur's Department of Mycology and with the Microbiology and the Food Chain Division at the French National Research Institute for Agriculture, Food and Environment (INRAE, USC 2019).
An approach for identifying other relevant genes, potential targets for antifungal therapy
To help understand the function of the CSA6 gene, the scientists used cell imaging techniques. "By tagging the Csa6 protein with a fluorescent protein, our colleagues at JNCASR were able to observe its localization in the nucleus, in the centrosome, the structure that separates chromosomes into the two cells during mitosis," continues Christophe d'Enfert.
The scientists also noticed that this protein was selectively present in yeast species related to C. albicans, like Candida dubliniensis, Candida tropicalis and Candida parapsilosis.
Aberrant, contrasting mitotic spindle structures after overexpression (left) and depletion (right) of Csa6 in Candida albicans cells with elongated buds. |
The ongoing study is the first to use gene overexpression screening at such a large scale in this human fungal pathogen. The findings determine and elucidate the functions of a new regulator of genome stability that is exclusively present in a group of yeasts including several medically relevant human fungal pathogens, while offering a systematic approach for identifying other genes.
The proteins identified represent potential new targets for antifungal therapy at a time when the development of drug targets to tackle pathogenic yeasts is urgently needed. Resistance to existing therapies is emerging in fungi in the genus Candida, which is responsible for often fatal infections in fragile patients – for example those in intensive care, with cancer or undergoing immunosuppressive therapy.
This study received funding from the Indo-French Centre for the Promotion of Advanced Research (IFCPAR/CEFIPRA). This financial support enabled the French and Indian groups to pool their expertise in functional genomics and the genome dynamics of human fungal pathogens.
Dr. Priya Jaitly, lead author of the article, is a former PhD student at the Jawaharlal Nehru Centre for Advanced Scientific Research (JNCASR) in the team led by Professor Kaustuv Sanyal, Head of the Molecular Mycology Laboratory and joint last author of the study. Dr. Priya Jaitly came to the Institut Pasteur in Paris to take part in this collaborative research.
Source
A phylogenetically-restricted essential cell cycle progression factor in the human pathogen Candida albicans, Nature Communications, July 23, 2022
Priya Jaitly1 , Mélanie Legrand2, Abhijit Das1,5, Tejas Patel1,5, Murielle Chauvel2, Corinne Maufrais3, Christophe d’Enfert2 & Kaustuv Sanyal1,4
1 Molecular Mycology Laboratory, Molecular Biology and Genetics Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore, India.
2 Institut Pasteur, Université Paris Cité, INRAE, USC2019, Fungal Biology and Pathogenicity Unit, F-75015 Paris, France.
3 Institut Pasteur, Université Paris Cité, Bioinformatics and Biostatistics Hub, F-75015 Paris, France.
4 Osaka University, Suita, Osaka, Japan.
5 These authors contributed equally