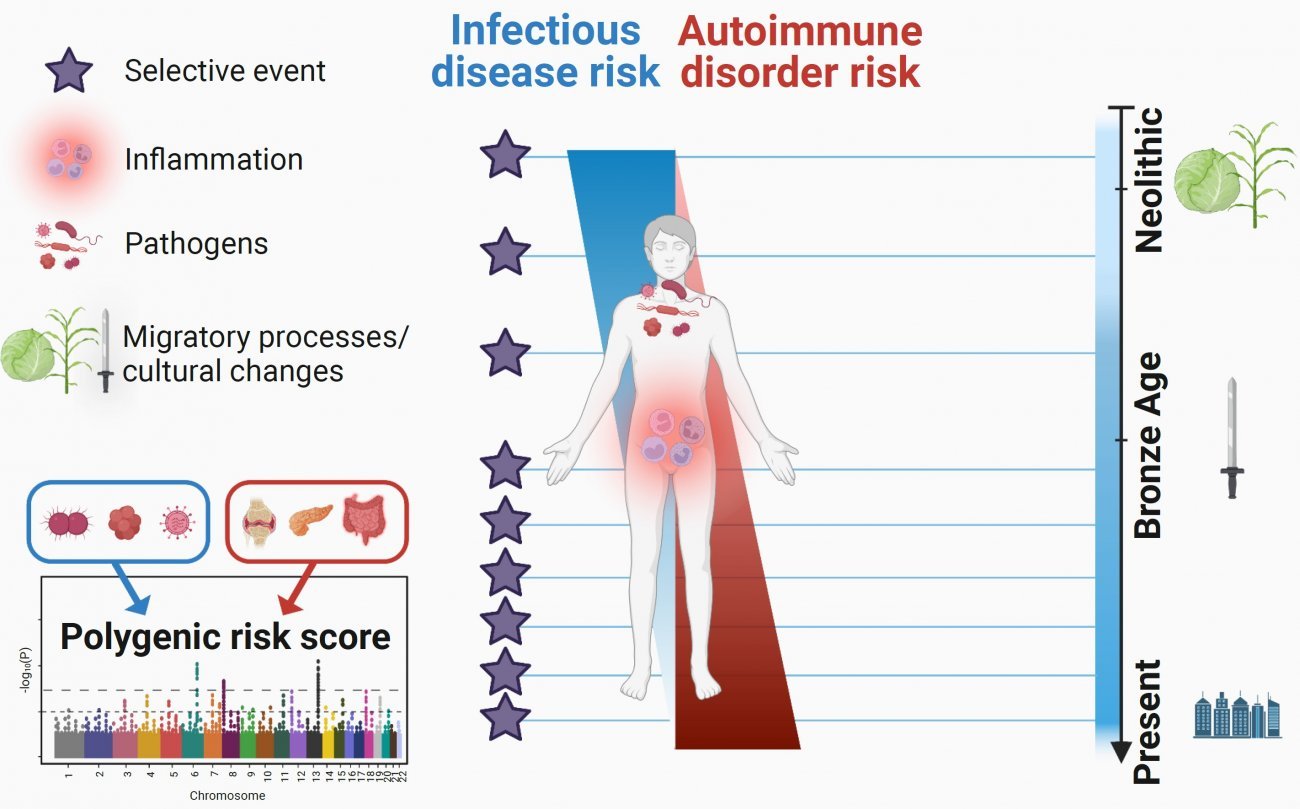
Scientists from the Institut Pasteur, Université Paris Cité, the CNRS and the Collège de France have used paleogenomics to trace 10,000 years of human immune system evolution. They analyzed the genomes of more than 2,800 individuals who lived in Europe over the past ten millennia. They were able to date the increase in frequency of most of the mutations that are advantageous in defending against pathogens to after the Bronze Age, 4,500 years ago. The scientists also observed that mutations conferring a higher risk of developing inflammatory disorders have become more frequent over the past 10,000 years. These enlightening results on the effects of natural selection on immunity genes were published in the journal Cell Genomics on January 13, 2023.
In the 1950s, the geneticist J.B.S. Haldane attributed the maintenance or persistence of the mutation responsible for anomalies in red blood cells commonly observed in Africa to the protection these anomalies provided against malaria, an endemic infection that claims millions of lives. This theory suggested that pathogens are among the strongest selective pressures faced by humans. Several population genetics studies subsequently confirmed the theory. But major questions remained, especially regarding the specific epochs during which the selective pressures exerted by pathogens on human populations were strongest and their impact on the present-day risk of developing inflammatory or autoimmune disorders.
To address these questions, scientists from the Institut Pasteur, Université Paris Cité, the CNRS and the Collège de France, in collaboration with the Imagine Institute and The Rockefeller University (United States), adopted an approach based on paleogenomics. This discipline, which studies the DNA from fossil remains, has led to major discoveries about the history and evolution of humans and human diseases, as illustrated by the decision to award the 2022 Nobel Prize in Physiology or Medicine to the paleogeneticist Svante Pääbo. In the study led by the Institut Pasteur, published on January 13 in the journal Cell Genomics, the scientists analyzed the variability of the genomes of more than 2,800 individuals who lived in Europe over the past ten millennia – a period covering the Neolithic, the Bronze Age, the Iron Age, the Middle Ages and the present.
By reconstituting the evolution over time of hundreds of thousands of genetic mutations, the scientists initially identified mutations that rapidly increased in frequency in Europe, indicating that they were advantageous. These mutations that evolved under "positive" natural selection are mainly located in 89 genes enriched in functions relating to the innate immune response, including especially the OAS genes – which are responsible for antiviral activity – and the gene responsible for the ABO blood group system. Surprisingly, most of these positive selection events, which demonstrate a genetic adaptation to the pathogenic environment, began recently, from the start of the Bronze Age, around 4,500 years ago. The scientists explain this "acceleration" in adaptation by the growth in the human population during this period and/or by strong selective pressures exerted by pathogens in the Bronze Age, probably linked to the spread of severe infectious diseases such as plague.
At the same time, the scientists also looked at the opposite situation, in other words, mutations whose frequency fell significantly over the past ten millennia. These mutations are probably subject to "negative" selection because they increase the risk of disease. They noted that once again, these selection events mainly began in the Bronze Age. Many of these disadvantageous mutations were also located in genes associated with the innate immune response, such as TYK2, LPB, TLR3 and IL23R, and have been confirmed in experimental research to have a deleterious effect in terms of infectious disease risk. The results emphasize the value of adopting an evolutionary approach in research on genetic susceptibility to infectious diseases.
Finally, the scientists explored the theory that the selection exerted by pathogens in the past gave an advantage to alleles conferring resistance to infectious diseases, but that in turn these alleles have increased the present-day risk of autoimmune or inflammatory disorders. They investigated the few thousand mutations known to increase susceptibility firstly to tuberculosis, hepatitis, HIV or COVID-19, and secondly to rheumatoid arthritis, systemic lupus erythematosus or inflammatory bowel disease. By looking at the evolution of these mutations over time, they observed that those associated with an increased risk of inflammatory disorders – including Crohn's disease – became more frequent over the past 10,000 years, while the frequency of those associated with a risk of developing infectious diseases decreased. "These results suggest that the risk of inflammatory disorders has increased in Europeans since the Neolithic period because of a positive selection of mutations improving resistance to infectious diseases," explains Lluis Quintana-Murci, director of the study and Head of the Human Evolutionary Genetics Unit (Institut Pasteur/CNRS Evolutionary Genomics, Modeling and Health Unit/Université Paris Cité).
The results of the study, which harnessed the huge potential of paleogenomics, show that natural selection has targeted human immunity genes over the past ten millennia in Europe, especially since the start of the Bronze Age, and contributed to present-day disparities in terms of the risk of infectious and inflammatory diseases.
As well as the institutions mentioned above, this research was supported by the French Foundation for Medical Research (FRM), the Allianz-Institut de France Foundation and the Fondation de France.
Explanatory diagram. © Gaspard Kerner, Institut Pasteur
Source
Genetic adaptation to pathogens and increased risk of inflammatory disorders in post-Neolithic Europe, Cell Genomics, January 13, 2023
Gaspard Kerner,1* Anna-Lena Neehus,2,3 Quentin Philippot,2,3 Jonathan Bohlen,2,3 Darawan Rinchai,4 Nacim Kerrouche,4 Anne Puel,2,3 Shen-Ying Zhang,2,3,4 Stéphanie Boisson-Dupuis,2,3,4 Laurent Abel,2,3,4 Jean-Laurent Casanova,2,3,4,5,6 Etienne Patin,1,8 Guillaume Laval,1,8 and Lluis Quintana-Murci,1,7,8,9,*
1Institut Pasteur, Université Paris Cité, CNRS UMR2000, Human Evolutionary Genetics Unit, F-75015 Paris, France
2Laboratory of Human Genetics of Infectious Diseases, INSERM UMR 1163, Necker Hospital for Sick Children, 75015 Paris, France.
3University Paris Cité, Imagine Institute, 75015 Paris, France.
4St. Giles Laboratory of Human Genetics of Infectious Diseases, The Rockefeller University, New York, NY 10065, United States.
5Howard Hughes Medical Institute, New York, NY 10065, United States
6Department of Pediatrics, Necker Hospital for Sick Children, 75015 Paris, France
7Collège de France, Chair of Human Genomics and Evolution, F-75005 Paris, France
8Senior author
9Lead contact
*Corresponding author





